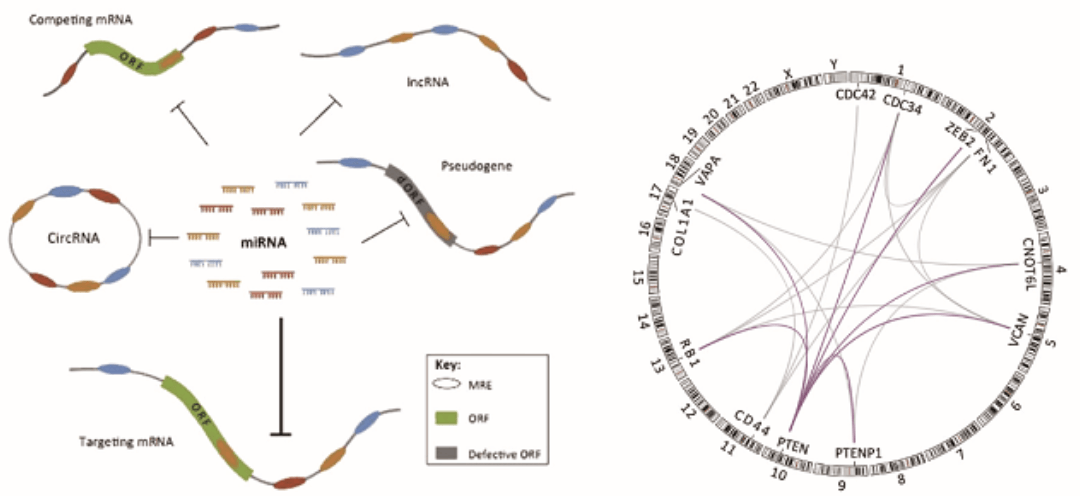
 Figure 1. Schematic diagram of ceRNA mechanism(Wang Y, et al. Trends Genet, 2016)
Figure 1. Schematic diagram of ceRNA mechanism(Wang Y, et al. Trends Genet, 2016)
In 2011, a landmark article "A ceRNA Hypothesis: The Rosseta Stone of a hidden RNA language?" was published in the journal CELL. The author put forward the "competitive Endogenous RNA (ceRNA) hypothesis" for the first time and proposed a "new language" for the interaction of RNA molecules in the ceRNA transcriptome. The theoretical core of the hypothesis is: the existence of ceRNA, these ceRNA molecules (mRNA, LncRNA and pseudogenes) can compete with the same miRNA through the MicroRNA Response Element (MRE) to regulate the expression level of each other. The proposal of this hypothesis opens a new door for transcriptome research.
CeRNA has attracted much academic attention in recent years. It represents a new regulation mode of gene expression. Compared with miRNA regulatory network, ceRNA regulatory network is more refined and complex, involving more RNA molecules, including mRNA, pseudogenes encoding genes, long-chain non-coding RNA, miRNA and so on. It provides a new perspective for researchers to study transcriptome, which is helpful to explain some biological phenomena more comprehensively and deeply. It is of great biological significance to study the regulatory mechanism of tumor ceRNA, and it is of great significance for later disease diagnosis and treatment, drug development and so on.
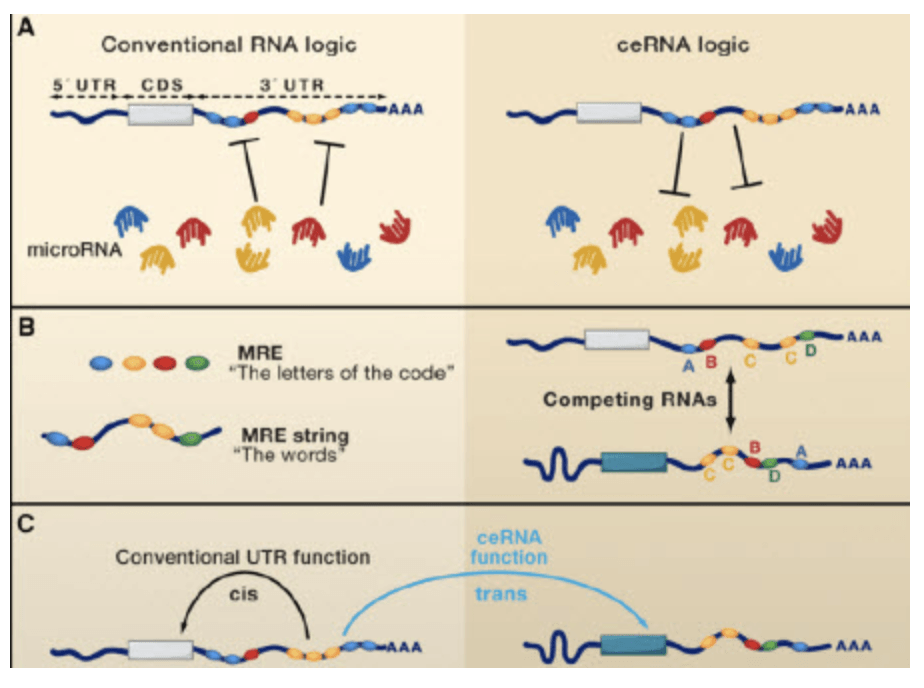 Figure 2. mRNAs as Regulators of miRNAs (L Slamena, et al. Cell. 2011)
Figure 2. mRNAs as Regulators of miRNAs (L Slamena, et al. Cell. 2011)
Features of ceRNA:
A) ceRNA has the same miRNA binding site.
B) ceRNA is regulated by miRNA.
There is a mutual regulation relationship between c) ceRNA, and the change trend of expression level is the same.
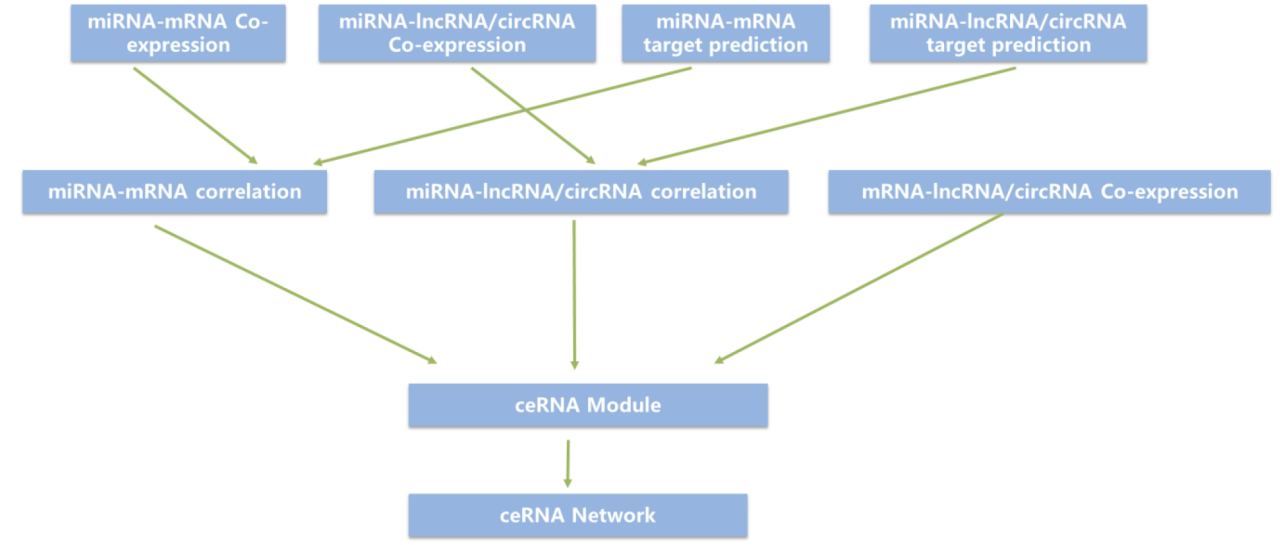
Overview of method
1. The expression information of mRNA, lncRNA, circRNA and miRNA in the tumor was obtained by sequencing, and the concerned RNA was screened by differential screening.
2. The target genes of miRNA, mRNA and lncRNA, that is, miRNA-mRNA and miRNA-lncRNA, were predicted by Miranda software. Similarly, the expression of RNA was used to predict the micorRNA-target relationship pairs composed of miRNA, mRNA and lncRNA, and the corrcoef function in python numpy library was used to screen the relationship pairs with high correlation. Combining the results of the two predictions, we take the intersection to improve the accuracy of the results.
3. The ceRNA_score principle (introduced below) is used to predict the formation of lncRNA-mRNA regulatory relationship pairs in ceRNA,. At the same time, the expression value was used to predict lncRNA-mRNA relationship pairs.Combining the results of the two-step analysis, we get our final predicted ceRNA regulatory relationship pair by taking the intersection of the results of the two-step analysis.
4.lncRNA functional annotations (KEGG, GO): the lncRNA here regulates the expression and degradation of mRNA by competing for limited miRNA, which is called ceRNA. LncRNA and mRNA communicate through a shared miRNA, so they have similar functions. Hypergeometric distribution test was used to calculate the enrichment significance of these mRNA regulated by lncRNA in GO or KEGG items. As a function comment of lncRNA.
5. The ternary relationship of lncRNA-miRNA-mRNA is shown in the form of a network diagram (as shown in the following figure). Cytoscape (version 3.4.0) software is used here to draw the relationship network diagram.
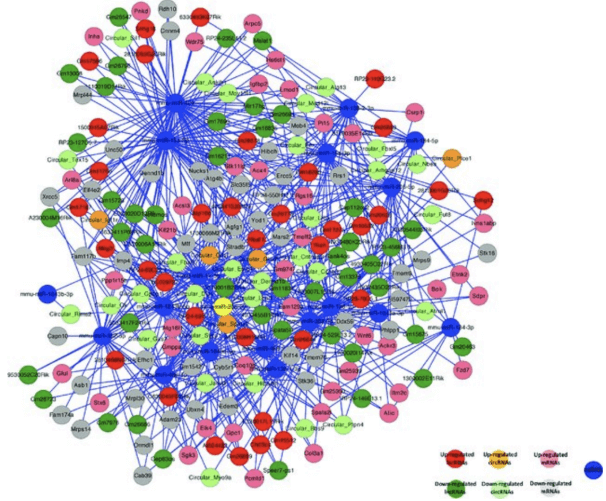 Figure 3. Competing endogenous RNA network in germline stem cells (Li X, et al. Oncotarget. 2011)
Figure 3. Competing endogenous RNA network in germline stem cells (Li X, et al. Oncotarget. 2011)
Our advantage
- A team of experts with decades of experience
- Powerful Bioinformatics Analysis Technology
- Joint analysis, mutual confirmation, enhance the persuasive power of the results
- To provide a comprehensive basis for the realization of accurate medical treatment
- High quality service, high level of experiment, reliable analysis
Sample type.
Cell, tissue, body fluid, serum, plasma, whole blood, total RNA, etc.
It is recommended that the starting dose of total RNA should be 60 μg, the minimum 30 μg, and the concentration ≥ 400 ng/ μL.
Sampling Kit: we provide our customers with a complete sampling kit, including protein and RNA separation kits, as well as tools for storing samples.
Deliverables: raw sequencing data, pruning and stitching sequences, quality control report results, statistics and bioinformatics reports you specify, visual pictures.
Reference
1. Li X, Ao J, Wu J.(2017). "Systematic identification and comparison of expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in mouse germline stem cell. Oncotarget, 8(16): 26573.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.
Related Services:

 Figure 1. Schematic diagram of ceRNA mechanism(Wang Y, et al. Trends Genet, 2016)
Figure 1. Schematic diagram of ceRNA mechanism(Wang Y, et al. Trends Genet, 2016) Figure 2. mRNAs as Regulators of miRNAs (L Slamena, et al. Cell. 2011)
Figure 2. mRNAs as Regulators of miRNAs (L Slamena, et al. Cell. 2011)
 Figure 3. Competing endogenous RNA network in germline stem cells (Li X, et al. Oncotarget. 2011)
Figure 3. Competing endogenous RNA network in germline stem cells (Li X, et al. Oncotarget. 2011)