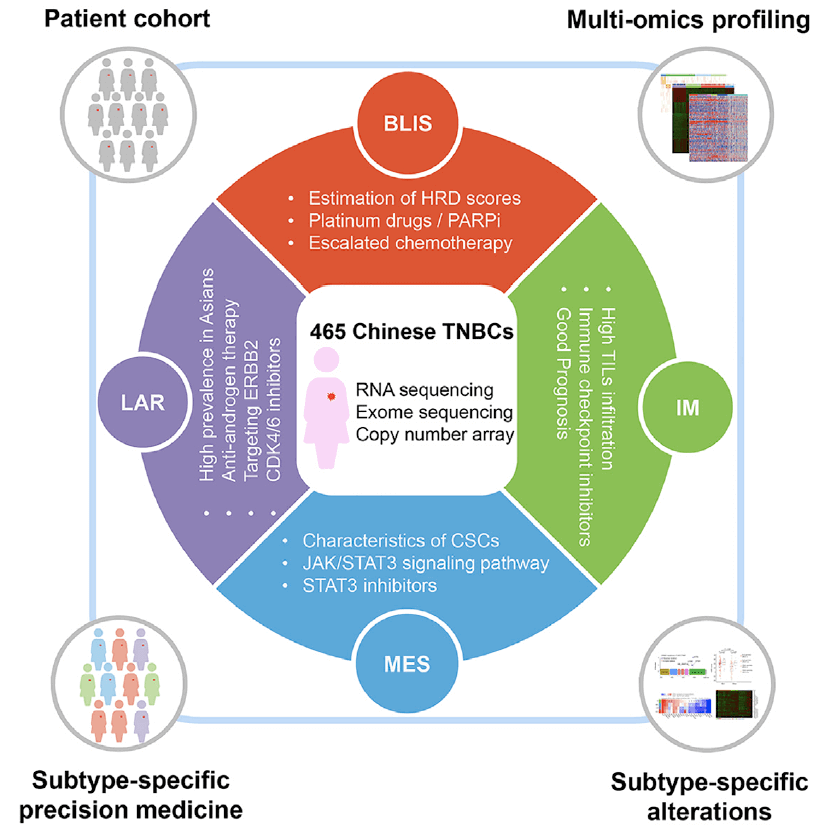
 Three negative breast cancer molecular typing(YiZhou Jiang, et al. 2019).
Three negative breast cancer molecular typing(YiZhou Jiang, et al. 2019).
Breast cancer is a "big family", which can be divided into four major subtypes: luminal A, luminal B, HER-positive and triple negative. Among them, triple negative breast cancer is the most difficult to treat. Cancer Cell recently published important results of breast cancer research online.In this study, 465 Chinese patients with primary triple-negative breast cancer (TNBC) were taken as the research object. Tumor tissue and adjacent tissues were collected, and WES, RNA-seq and cnv chip technology were used to obtain mutation sites, differentially expressed gene analysis and cnv site for tumor typing.In this study, 419 TNBC samples with mutations or copy number variations were sorted by mRNA subtypes and mutation profiles, and the genome map of triple-negative breast cancer was drawn. The most significant cancer-related mutation observed in this cohort was the TP53 mutation ( Found in 74% of tumors), followed by PIK3CA (18%), KMT2C (7%), and PTEN (6%) mutations (Figures 1A and 1B). By evaluating the relationship between somatic cell copy number changes (CNAS) and the reported changes in oncogenes and tumor suppressor genes, MYC was found to be the most frequently affected gene for CNAS in recipient cells (Figure 1).
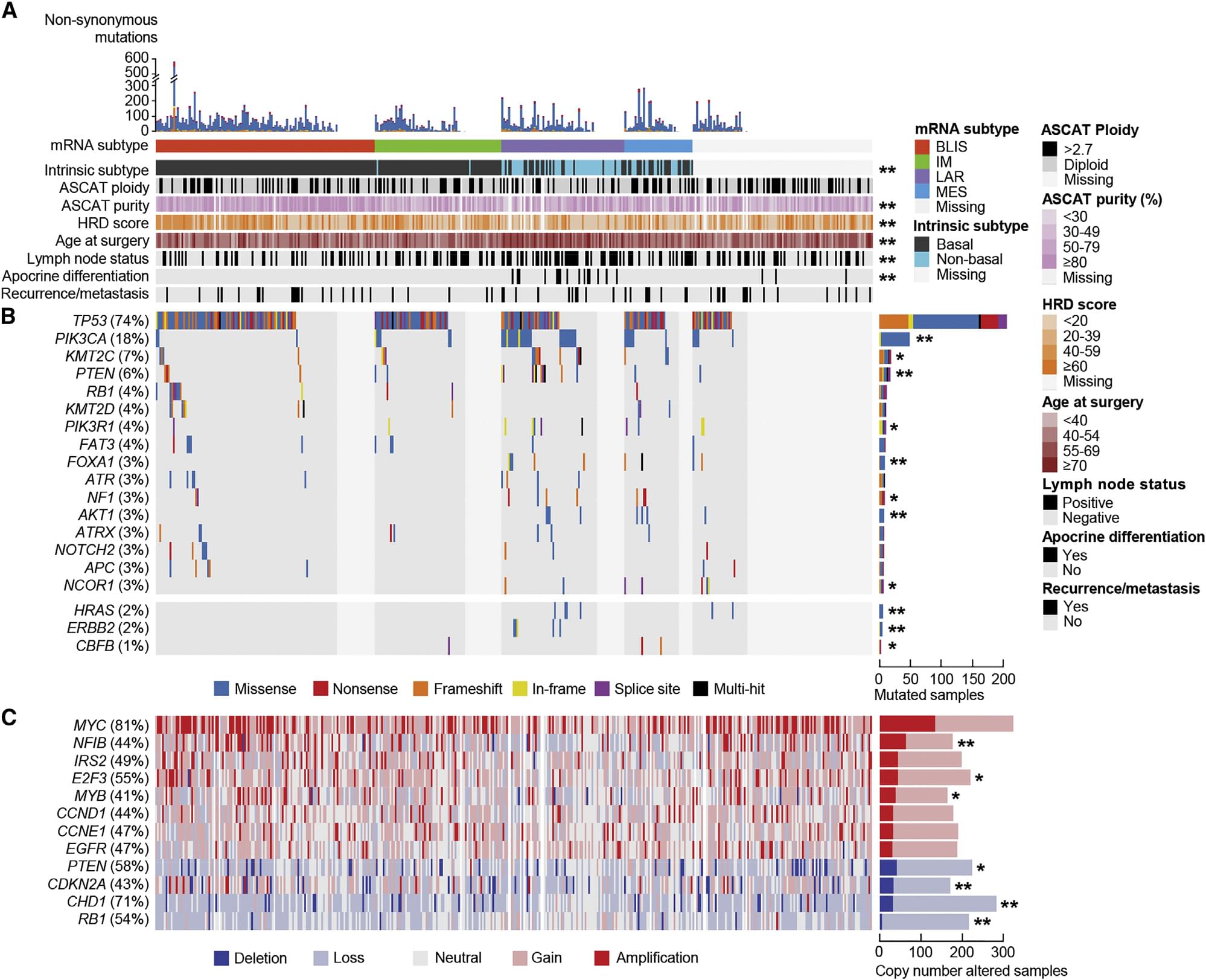 Figure 1. TNBC genome map of Chinese population
Figure 1. TNBC genome map of Chinese population
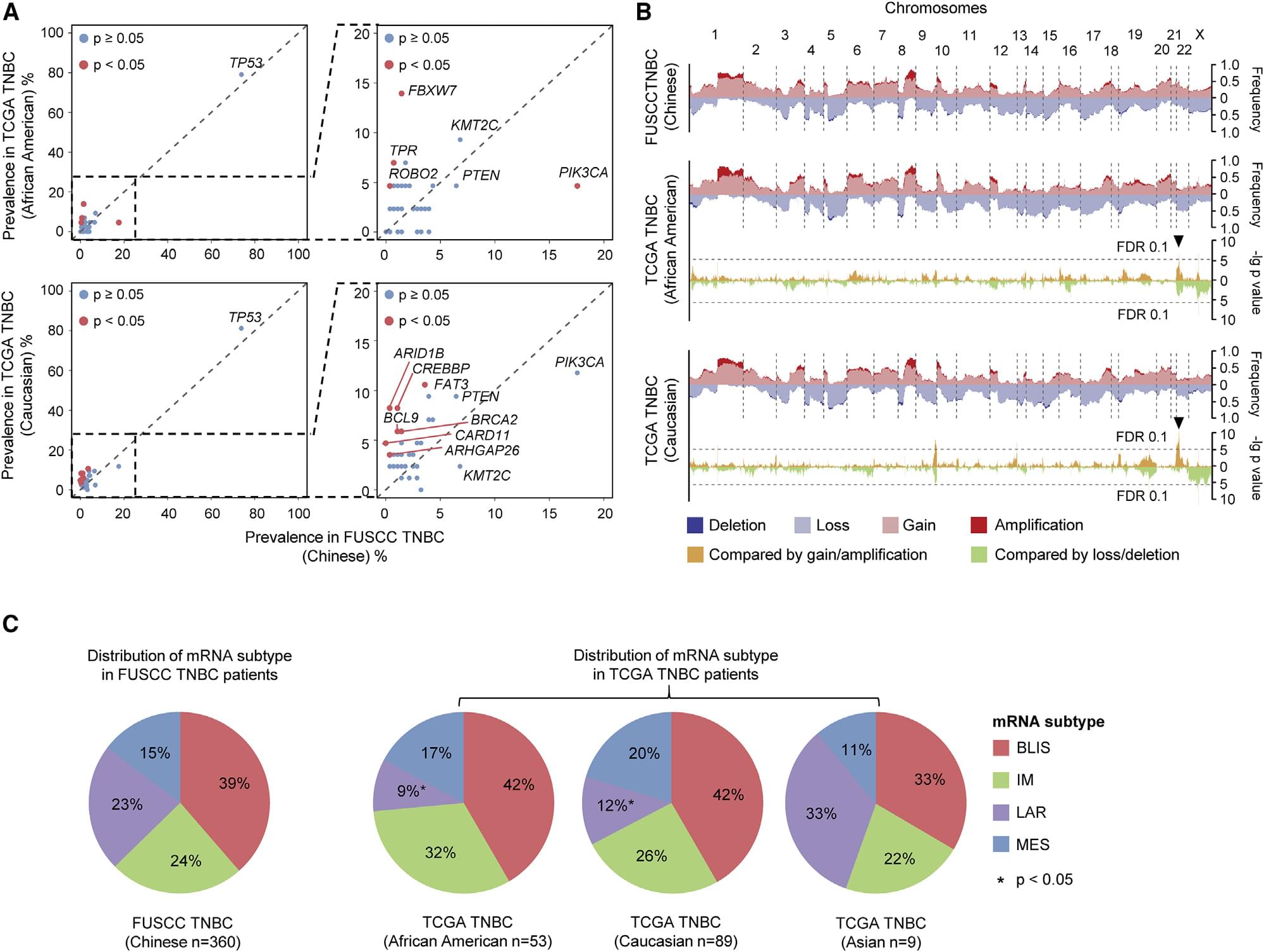 Figure 2. Chinese TNBC population-specific genomic variation.(YiZhou Jiang, et al. 2019).
Figure 2. Chinese TNBC population-specific genomic variation.(YiZhou Jiang, et al. 2019).
The study first announced a group of Chinese people with triple-negative breast cancer-specific gene mutations, such as PIK3CA gene mutations in China's population of triple-negative breast cancer patients significantly higher than the US data. The PIK3CA mutation rate of Chinese TNBCs is higher than that of TCGA TNBC (18% and 10%, respectively). The main reason may be the difference between African and Chinese TNBCs (5% and 18%, respectively, Figure 2A). In terms of CNA differences, the frequency of somatic cell copy number increase on Chinese chromosome 22q11 is higher than that of TCGA TNBC (Chinese: TCGA African American: TCGA Caucasian = 44%: 19%: 15%, Figure 2B) Compared with TCGA TNBC patients, the prevalence of LAR subtypes in Chinese is higher (23% are Chinese, 9% are TCGA African Americans, and TCGA Caucasians are 12%, as shown in Figure 2C).
Subtyping Chinese TNBC based on multiomics data
Based on more than 2,000 differentially expressed K-means clustering coding genes, TNBC was divided into 4 separate subtypes (Figure 3A), respectively: intraluminal androgen receptor (LAR) subtype (23%); immunity Modulation (IM) subtype (24%) with high immune cell signal and cytokine signal gene expression; basal-like immunosuppressive (BLIS) subtype (39%), which is characterized by up-regulation of the cell cycle, activation of DNA repair and Down-regulated immunity; Mesenchymal-like (MES) subtypes (15%), which are enriched in the breast stem cell pathway. Using K-means clustering and consensus clustering similar to mRNA clustering, six clusters based on CNA peaks were identified (Figure 3B). In addition, based on the classification of cancer mutation characteristics, four subtypes were identified (Figure 3C).
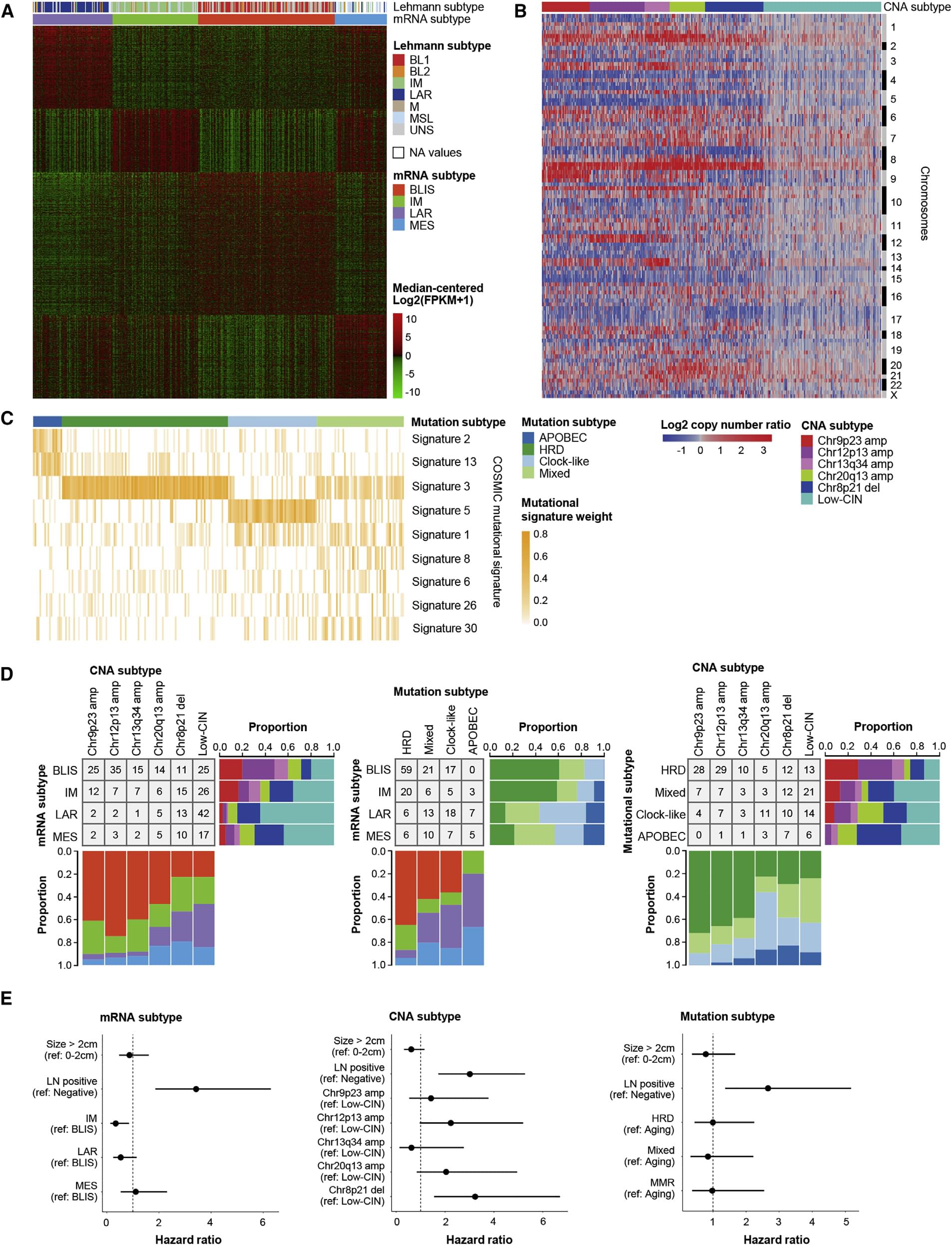 Figure 3. Subtyping of TNBC in China using multiomics data.
Figure 3. Subtyping of TNBC in China using multiomics data.
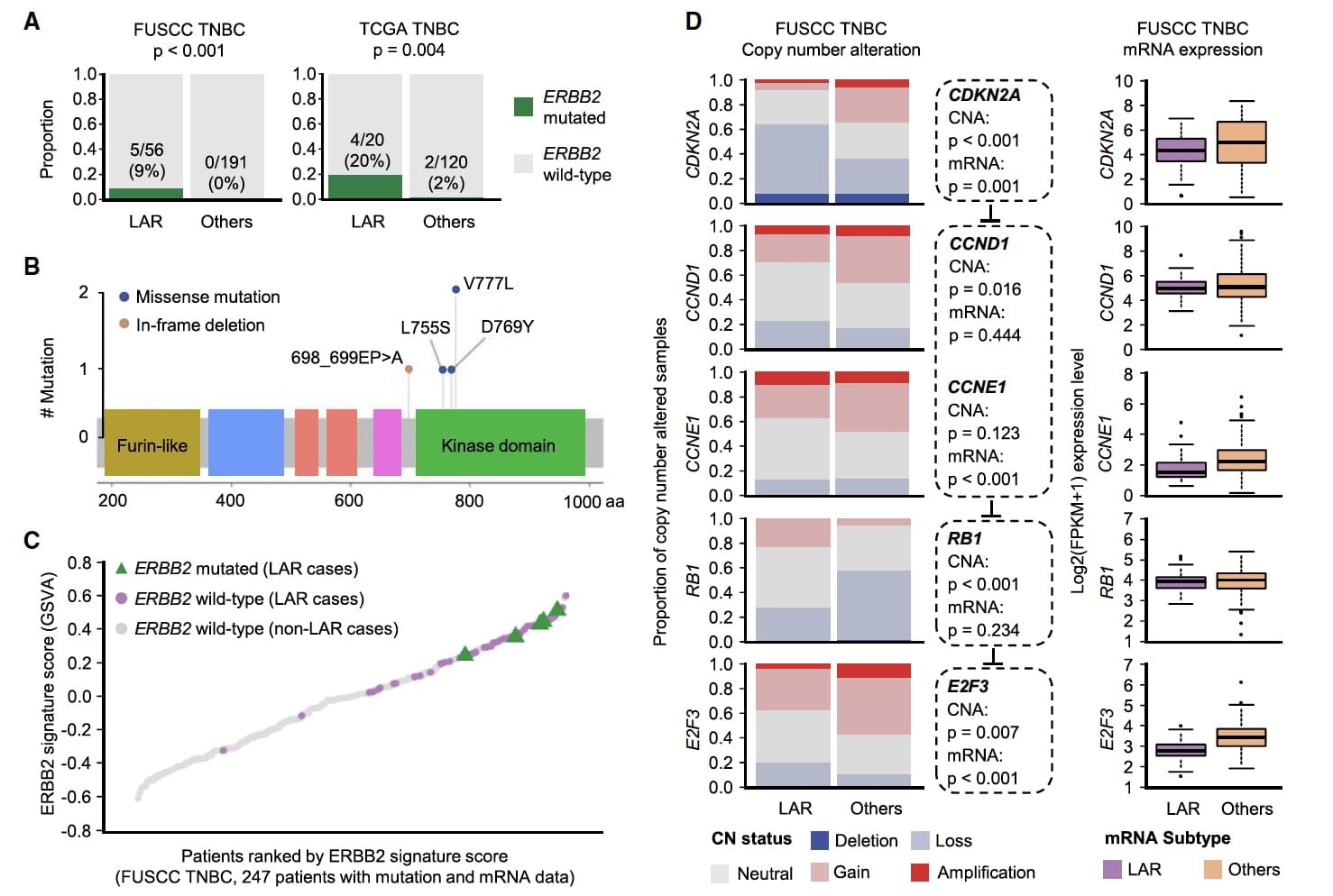 Figure 4. Activated ERBB2 and Cell-Cycle Signaling in the LAR Subtype
Figure 4. Activated ERBB2 and Cell-Cycle Signaling in the LAR Subtype
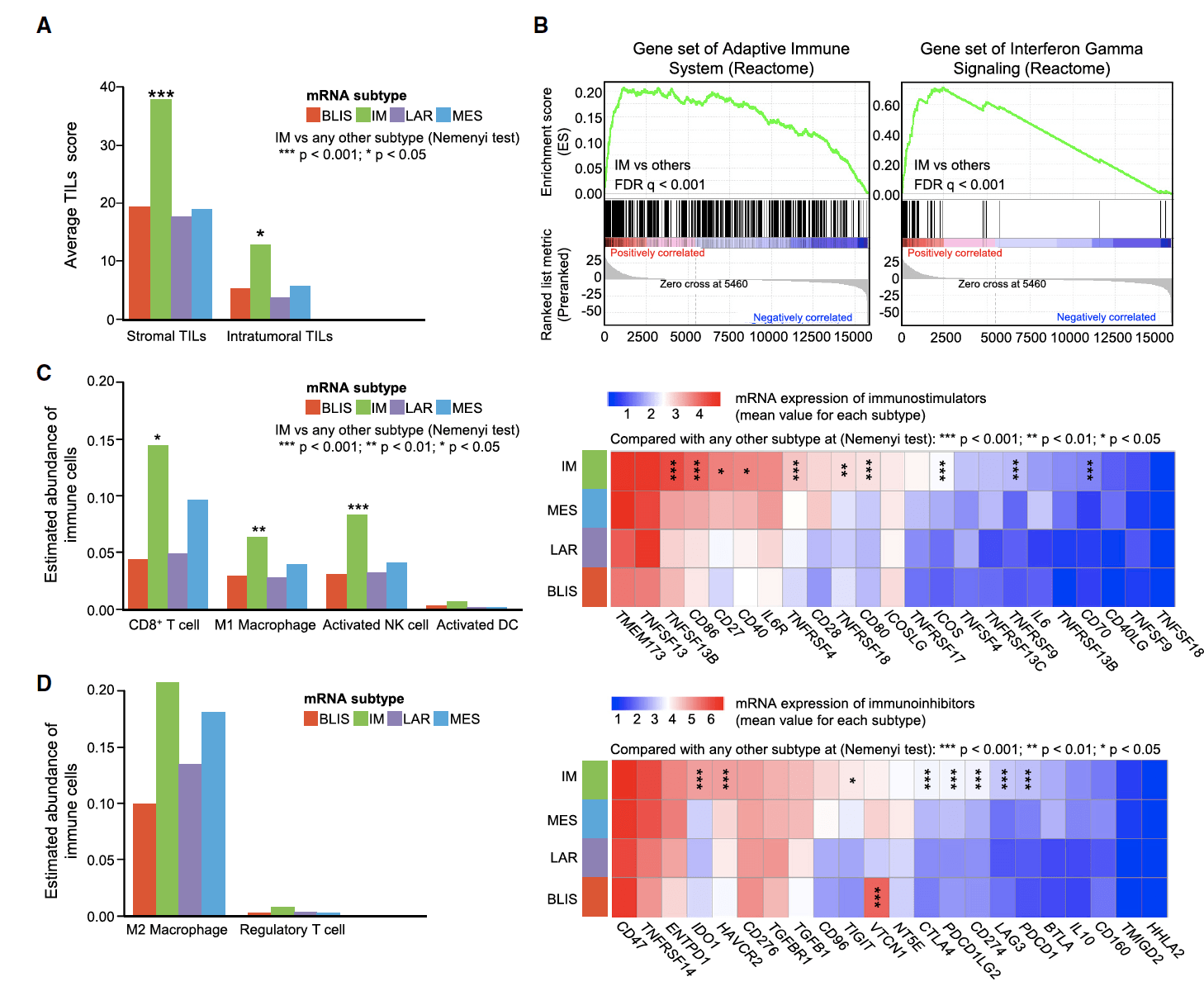 Figure 5. Potential Application of Immune Checkpoint Blockade in the IM Subtype(YiZhou Jiang, et al. 2019).
Figure 5. Potential Application of Immune Checkpoint Blockade in the IM Subtype(YiZhou Jiang, et al. 2019).
This study is the first internationally proposed triple-negative breast cancer classification standard based on a multidimensional big data system. To explore potential therapeutic targets for the four TNBC subtypes, the authors explored different genomic variations. Five of the LAR patients had ERBB 2 nonsynonymous mutations, and ERBB 2 mutations included activating mutations V777L (2 cases), D769Y (1 case), and L755S (1 case). Further research found that these types of site mutations not only played a key role in the activation of the ERBB 2 gene and ERBB 2 pathway, but also resistance to trastuzumab, lamatinib, and previously unreported E698_P699 delinsA (Figure 4B, 4C ). The deletion of Chr9p21 in the LAR subtype also affects the mutation of CDKN2A, a key gene that regulates the cell cycle. CDKN2A and E2F3 expression levels are significantly reduced in LAR patients (Figure 4D). Therefore, it is concluded that LAR TNBCs are potentially sensitive to CDK 4/6 inhibitors or other cell cycle inhibitors.The main conclusions of this study are the following four points: 1. Establishing genomic and transcriptome multi-omics maps of 465 primary primary TNBCs; 2. Discovering that Chinese TNBC cases show more PIK3CA mutations and LAR subtypes; TNBC is divided into four subtypes; 4. Potential therapeutic targets among the four TNBC subtypes are identified through multiomics studies.
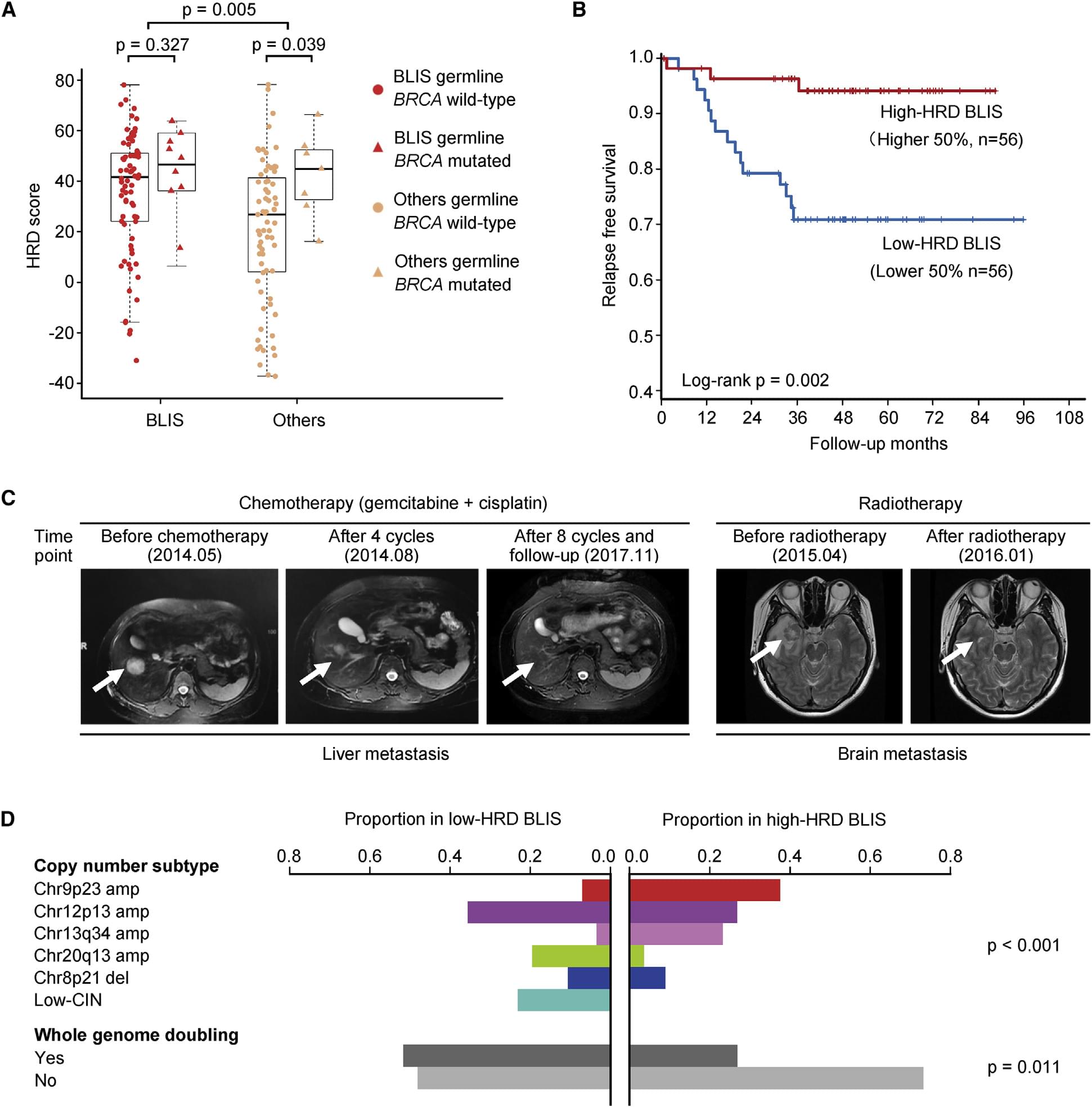 Figure 6. Further Classification of the BLIS Subtype According to HRD Score(YiZhou Jiang, et al. 2019).
Figure 6. Further Classification of the BLIS Subtype According to HRD Score(YiZhou Jiang, et al. 2019).
Multi-omics research can be described as a great tool for obtaining high-scoring articles in recent years. It not only promotes the research of pathogenic mechanisms, target screening, disease prevention and treatment, but also provides new ideas for basic disease research and precision medicine research. The combined analysis of genomics, transcriptomes, and epigenomics introduced by Anuo Gene can systematically and comprehensively explain the mechanism of carcinogenesis, and use multiomics data to jointly screen for key genes and related signal pathways of tumorigenesis to accurately locate key Genetic changes, multidimensional analysis of pathogenic mechanisms.Creative proteomics has an excellent team of experts to provide you with customized multi-group oncology-related experimental design and experimental solutions, so that you can experience the highest quality, the most advanced and the most satisfactory service.Help scientists in the field of cancer research to produce quick results and good results, so as to promote scientific and technological innovation. Creative proteomics, multi-layer assemblage customization service experts to help you with your scientific research!
Reference
1. YiZhou Jiang, Ding Ma, Chen Suo, et al. (2019)."Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies." Cancer Cell, 35(3): 428-440.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 Three negative breast cancer molecular typing(YiZhou Jiang, et al. 2019).
Three negative breast cancer molecular typing(YiZhou Jiang, et al. 2019). Figure 1. TNBC genome map of Chinese population
Figure 1. TNBC genome map of Chinese population Figure 2. Chinese TNBC population-specific genomic variation.(YiZhou Jiang, et al. 2019).
Figure 2. Chinese TNBC population-specific genomic variation.(YiZhou Jiang, et al. 2019). Figure 3. Subtyping of TNBC in China using multiomics data.
Figure 3. Subtyping of TNBC in China using multiomics data. Figure 4. Activated ERBB2 and Cell-Cycle Signaling in the LAR Subtype
Figure 4. Activated ERBB2 and Cell-Cycle Signaling in the LAR Subtype Figure 5. Potential Application of Immune Checkpoint Blockade in the IM Subtype(YiZhou Jiang, et al. 2019).
Figure 5. Potential Application of Immune Checkpoint Blockade in the IM Subtype(YiZhou Jiang, et al. 2019). Figure 6. Further Classification of the BLIS Subtype According to HRD Score(YiZhou Jiang, et al. 2019).
Figure 6. Further Classification of the BLIS Subtype According to HRD Score(YiZhou Jiang, et al. 2019).