Fishbein L and other researchers used oncology platforms to integrate and analyze the comprehensive molecular characterization of a rare tumor type-pheochromocytoma and paraganglioma (PCC/PGLs), genomics involves genome, transcriptome, miRNA, methylation, proteomics and other data for extensive joint analysis. Pheochromocytoma (PCC)and paraganglioma(PGL)are neuroendocrine tumors that originate from chromaffin cells and occur in adrenal medulla (PCC) and sympathetic ganglion or parasympathetic ganglion (PGL). The researchers collected and analyzed the PCC/PGL cohort of 173 patients through TCGA. In order to identify and characterize the genomic changes of PCC/PGL, tissue samples were analyzed by a variety of genome analysis methods.
Figure 1. Germline and Somatic Genome Alterations
Genomic features are in progress, with primary tumors (nasty 173) in the list; shadows indicate the effect of mutations on protein sequences. Significant somatic mutations (MutSig2,q<0.05) are indicated by an asterisk (*).(Fishbein L, et al. 2017)
Matching normaltissues and tumor specimens were analyzed for mutations by full exome sequencing, and SNP arrays were used for copy number analysis. Tumor specimens were also analyzed by mRNA sequencing, miRNA sequencing, DNA methylation arrays, and reversed-phase protein arrays for targeted proteome analysis. Our analytical strategy includes systematic interrogation through the platform to identify genomic changes in PCC / PGL, including germline mutations, somatic mutations, fusion genes and copy number changes.
Perform multi-platform integration and calculation analysis:
(1) Characterize broad molecular correlations with significant driver changes;
(2) Determine the PCC / PGL molecular subtype classification;
(3) identify the interrupted path;
(4) Identification of molecular identification factors for metastatic disease.
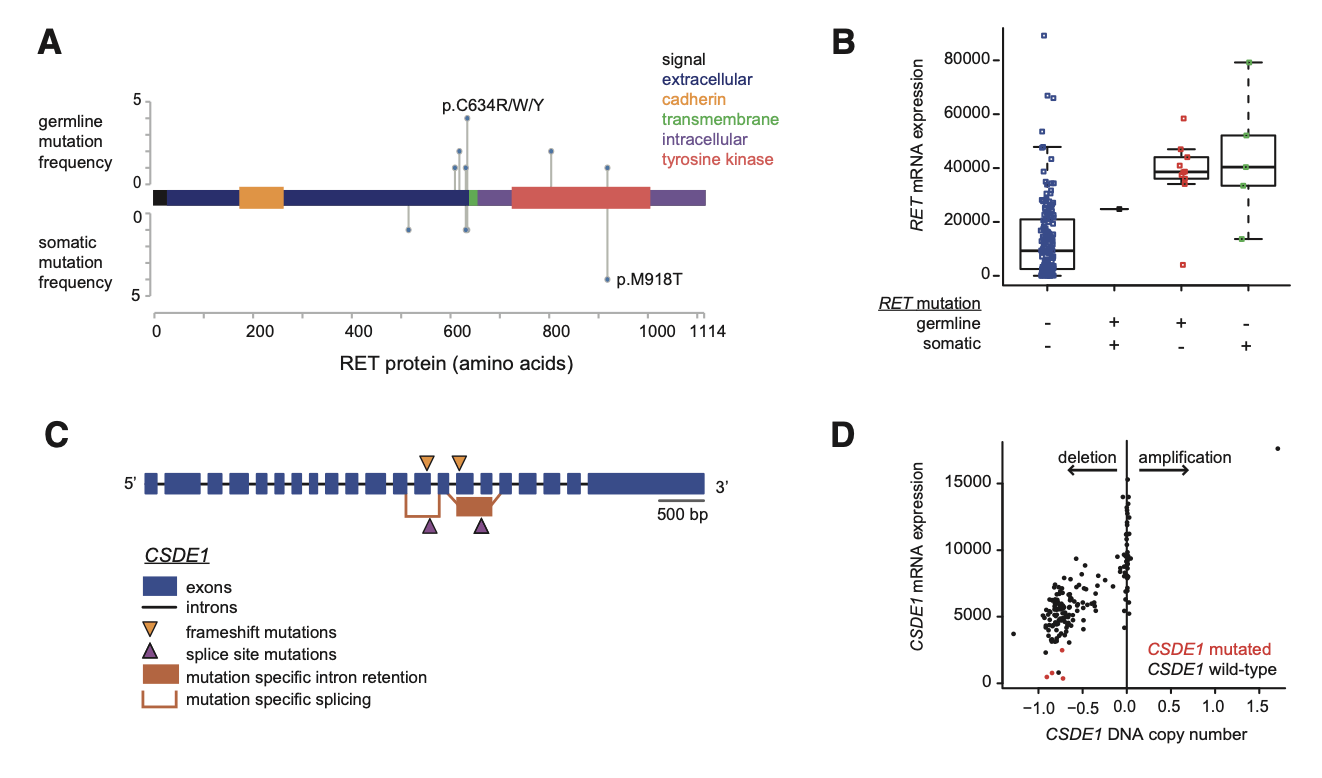 Figure2. Integrated Alterations in RET and in CSDE1(Fishbein L, et al. 2017)
Figure2. Integrated Alterations in RET and in CSDE1(Fishbein L, et al. 2017)
Known from Figure 2, (A) Location of somatic and germline RET mutations within the protein sequence.(B) RET mRNA expression in mutation-positive (+) and mutation-negative (-) tumors. The horizontal line of the boxplot indicates the 25th, 50th, and 75%, and the line extends to the farthest point, which is less than or equal to 1.5 times the interquartile range. The dots represent the primary tumor, and horizontal dithering has been added to help visualization.(C) Mutations within the structure of the CSDE1 gene.(D) Relationship between CSDE1 mRNA expression and CSDE1 DNA copy number, dots represent primary tumors.
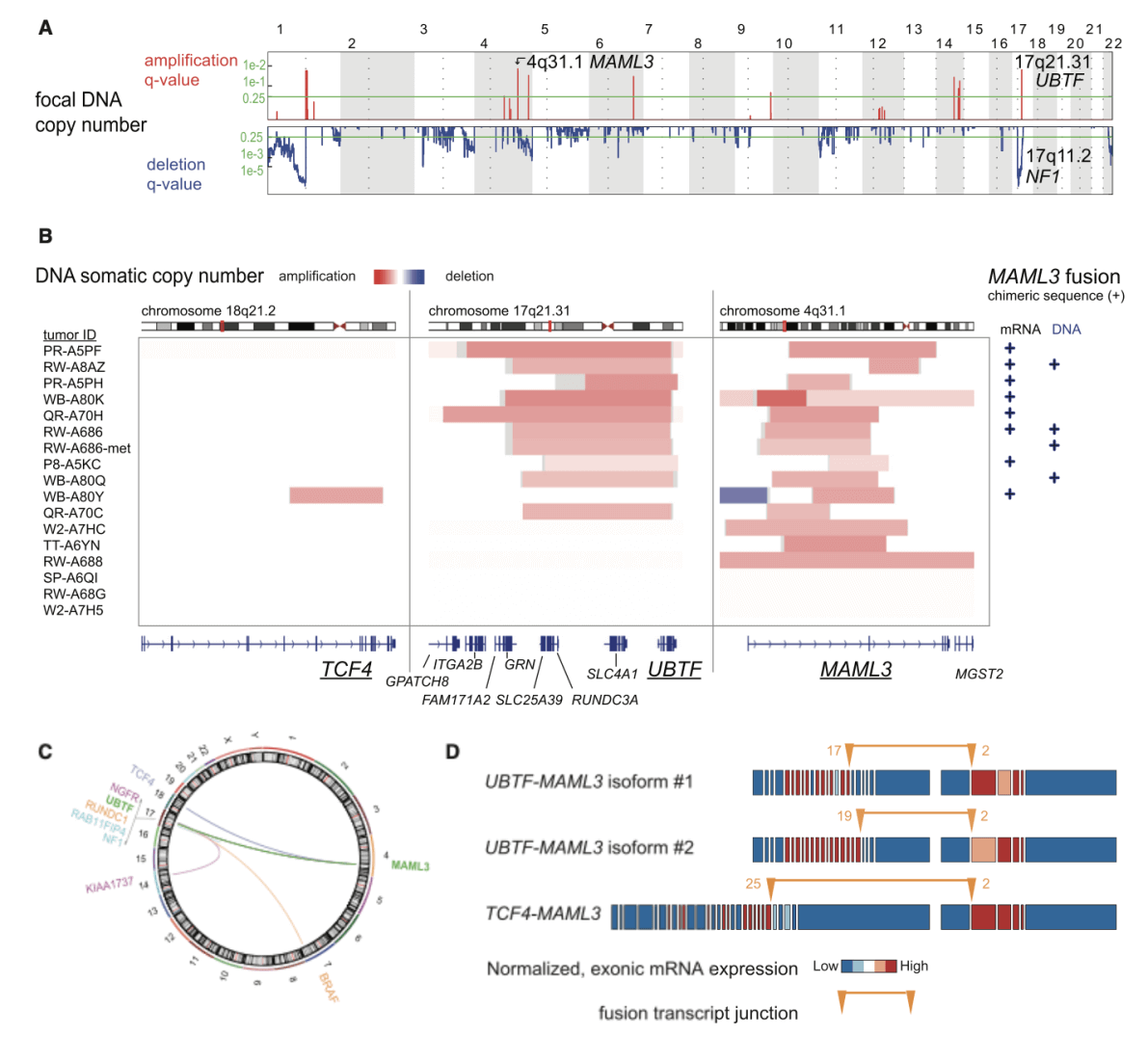
Figure 3. Molecular Correlates of MAML3 Fusion
(A) Probes for differential methylation in tumors by MAML3 fusion status.
(B) Log2 ratio of selected mRNA, miRNA and DNA methylation markers.
(C) The horizontal line of the boxplot indicates the 25th, 50th, and 75th percentiles, and the whiskers extend to the farthest point, which is less than or equal to 1.5 times the interquartile range. The dots represent the original tumor values, and horizontal dithering has been added to help visualization.(Fishbein L, et al. 2017)
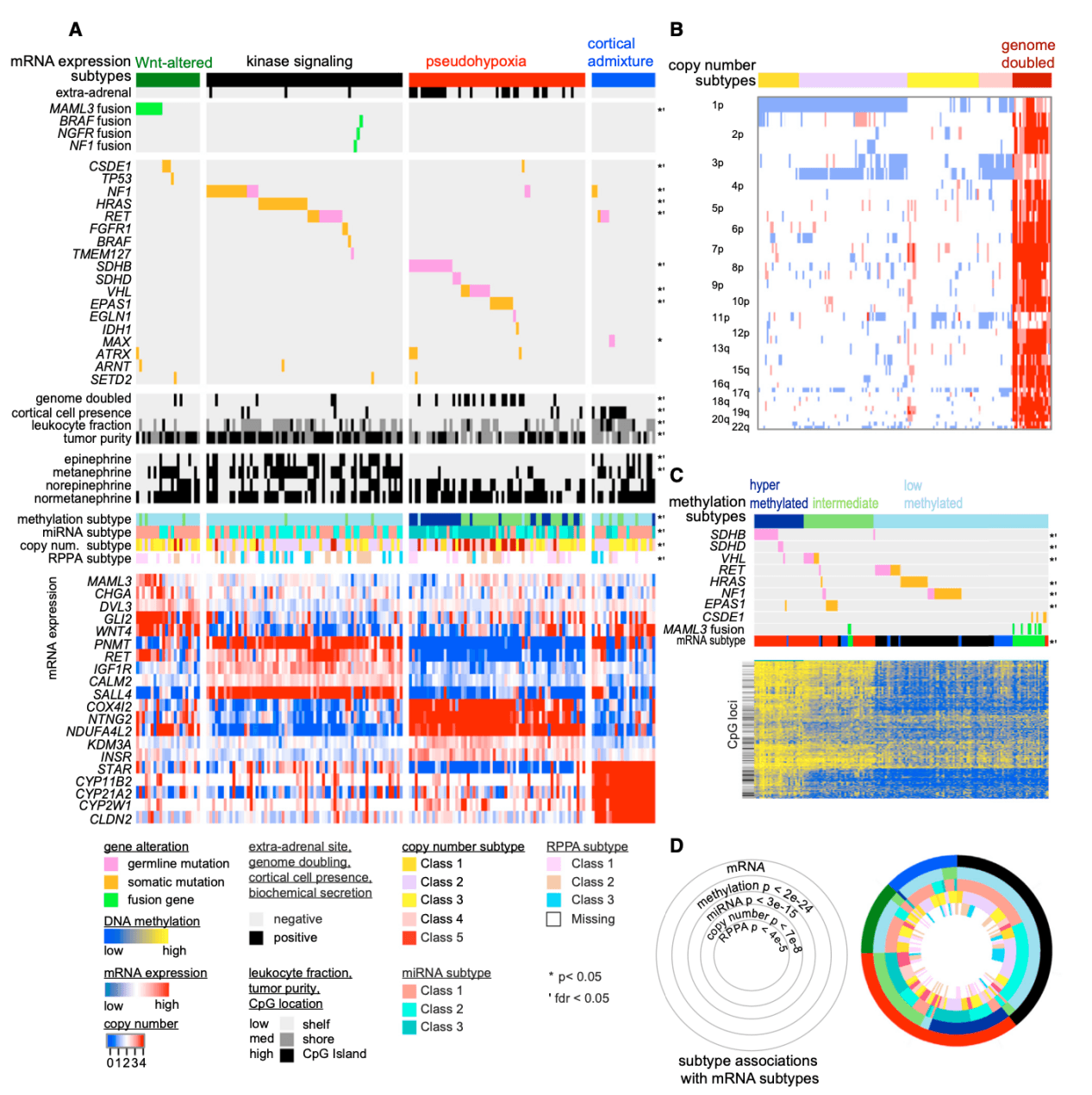
Studies have found that PCCs / PGLs new driver mutation gene CSDE1, CSDE1 mutations are often accompanied by significant gene deletion and down-regulation; tumors containing fusion genes related to MAML3, BRAF, NGFR and NF1 often show gene overexpression, hypermethylation or DNA amplification, etc .; PCCs / PGLs are divided into 4 subtypes based on mRNA data: kinase signal subtype, pseudo hypoxia subtype, Wnt mutant and cortical mixed type. Methylation, CNV, miRNA, and RPPA data are all related to this. The four subtypes are significantly related; in-depth analysis of each subtype and integration with clinical data provide a comprehensive theoretical basis for the realization of precision medical treatment of PCCs / PGLs.
Figure 4. Integrated Molecular Subtypes
(A) mRNA subtype. Primary tumors (n = 173) are shown in columns, and clinical and genomic features are shown in rows. Classification features were analyzed using Fisher's exact test; continuous features were analyzed using Kruskal-Wallis test. The selected differentially expressed genes are shown below each subtype.
(B) DNA copy number clustering. Primary tumors appear in the column (n = 173).
(C) DNA methylation clustering. Primary tumors (n = 173) appear in the column.
(D) A ring diagram showing cross-platform subtype associations. p tends to perform chi-square tests on platform subtypes and mRNA expression subtypes.(Fishbein L, et al. 2017)
This study integrates genome, transcriptome, miRNA, methylation, proteomics and other data for extensive joint analysis. PCCs/PGLs is divided into four molecular subtypes, and in-depth characteristic analysis is carried out. At the same time, it is integrated with clinical data, which provides a comprehensive theoretical basis for the realization of PCCs/PGLs precision medicine. Creative Proteomics has an excellent team of experts to provide you with customized multi-group oncology-related experimental design and experimental solutions, so that you can experience the highest quality, the most advanced and the most satisfactory service. Help scientists in the field of cancer research to produce quick results and good results, so as to promote scientific and technological innovation. Creative Proteomics, multi-layer assemblage customization service experts to help you with your scientific research!
Reference
1. Fishbein L, Leshchiner I, Walter V, et al. (2017). "Comprehensive molecular characterization of pheochromocytoma and paraganglioma." Cancer cell. 31(2): 181-193.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

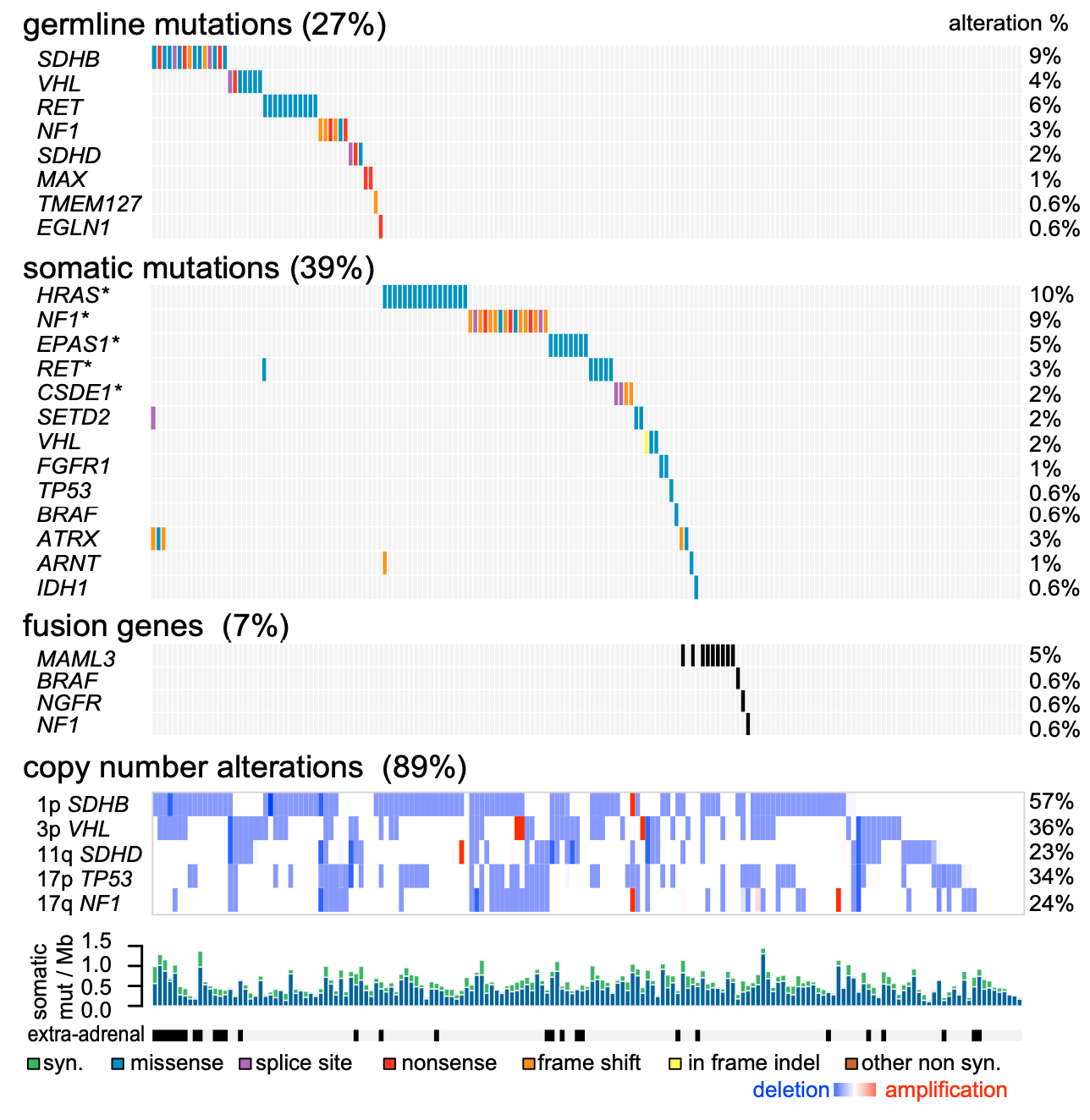
 Figure2. Integrated Alterations in RET and in CSDE1(Fishbein L, et al. 2017)
Figure2. Integrated Alterations in RET and in CSDE1(Fishbein L, et al. 2017)