Article highlights:
1. mutation-phosphorylation correlation suggests possible signaling interplays in EOGC
The correlation between gene mutation and protein phosphorylation suggests a possible signaling pathway effect in early-onset gastric cancer.
2. mRNA-protein correlation suggests genes with high association with patient survival
The mRNA-protein correlation suggests genes that are highly correlated with patient survival.
3. Integrated analysis of mRNA and protein data identified four subtypes
The integrated analysis of mRNA and protein data divides early gastric cancer into four subtypes.
4. Phosphorylation data provide cellular signaling pathways underlying the subtype
Protein phosphorylation data provide information on cellular signaling pathways for each subtype.
In an article on proteomics of early-onset gastric cancer published in Cancer cell in 2019, using tumor multi-group regulation mechanism, the researchers obtained tumor tissues and paired adjacent normal tissues from 80 patients, using exon sequencing (tumor tissues,); mRNA sequencing of peripheral blood monocytes (tumor tissues and adjacent normal tissues); The proteomics of early-onset gastric cancer was analyzed by whole proteome, phosphorylated proteome and N-glycosylated proteome (all aimed at tumor tissues and adjacent normal tissues).
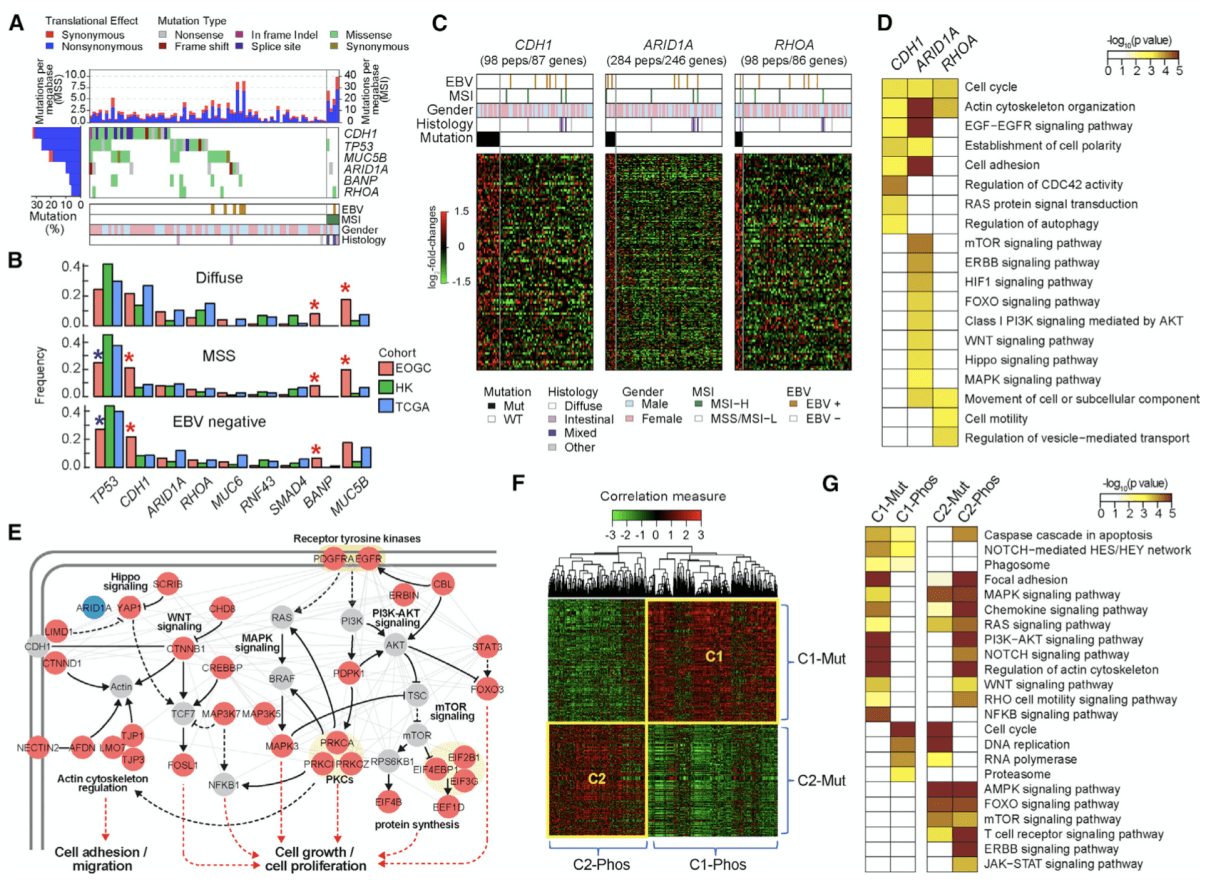 Figure 1. Proteogenomic Analysis of Nonsynonymous Somatic SNVs .(Mun DG, et al. 2019)
Figure 1. Proteogenomic Analysis of Nonsynonymous Somatic SNVs .(Mun DG, et al. 2019)
In this article, EOGC cohort identified 6 significant mutation genes, of which MUC5B and BANP were significant mutation genes that have not been reported before. The SNV frequencies of non-synonymous somatic cells in the three GC types (diffusive, microsatellite stable, and EBV negative) of EOGC and predecessor LOGC were compared. In diffuse gastric cancer, the mutation frequency of BANP and MUC5B in EOGC is higher than that in LOGC. In microsatellite stable, EBV-negative gastric cancer, the mutation rates of CDH1, BANP and MUC5B in EOGC were also higher than those in LOGC. Researchers analyzed the correlation between somatic SNV and protein phosphorylation levels, and the correlation between mRNA and protein log2-FoldChange, etc., EOGC cohort identified a total of 6 significant mutant genes, of which MUC5B and BANP were not previously reported Significantly mutated genes. The SNV frequencies of non-synonymous somatic cells in the three GC types (diffusive, microsatellite stable, and EBV negative) of EOGC and predecessor LOGC were compared. Of the four significant mutated genes common to the three cohort, phosphorylation levels were identified in samples containing these mutated genes. Among them, three mutant genes (CDH1, ARIDA, RHOA) showed mutation-phosphorylation correlation in 80 proteins, and these proteins were significantly enriched in processes and pathways related to cell adhesion and cell movement. CDH1 inactivated by mutations causes heritable diffuse gastric cancer. RHOA mutations have been reported to be common in diffuse gastric cancer and affect cell proliferation and the spread of gastric cancer. ARIDA also affects cell proliferation and differentiation.
Figure 2. Correlation between mRNA and Protein Abundance Changes(Mun DG, et al. 2019)
Next, the researchers selected 6803 genes, and their protein and mRNA abundances were more than 30% in both tumors and adjacent tissues. Of these genes, only 34.4% showed a significant positive (FDR <0.01) average correlation coefficient of 0.28; 1000 genes with strong correlation were related to DNA replication, PPAR signaling pathway, and amino acid metabolism. The 1,000 weakly related genes are related to spliceosome, mRNA supervision, and proteolysis. The authors first analyzed which genes mRNA-protein correlation was positively or negatively correlated with survival. Because tumor suppressor and proto-oncogene are positively or negatively correlated with survival, respectively, as shown in the figure below. Using significant mRNA-protein correlations, tumor suppressor genes and proto-oncogenes can be identified. The TCGA GC data set was used to analyze whether these genes were related to patient survival. For each gene, patients with the first 25% and last 25% of their mRNA expression were selected to analyze the survival differences between the two groups of patients. Genes with significant mRNA-protein correlation (FDR <0.1) showed greater survival differences. Among 2334 genes with significant mRNA-protein correlation, 217 genes were selected whose mRNA expression level was significantly positively or negatively correlated with survival. Genes positively related to survival and antitumor processes are related, genes negatively related to survival and cell invasion are related.Three ACRG GC mRNA datasets were used to try to find tumor suppressor genes or proto-oncogenes from these genes with significant mRNA-protein correlation. Three genes that are positively related to survival and 21 genes that are negatively related to survival were found. From the EOGC data set, two tumor suppressor genes and 15 proto-oncogenes were further selected. Two GC cell lines were used for siRNA transcription and gene knockout experiments to verify the reliability of the genes selected.
Figure 3. Subtypes of EOGC Identified by an Integrated Proteogenomic Analysis(Mun DG, et al. 2019)
Subsequently, theauthors integrated proteomics-genomics to type EOGC to examine whether the mRNA and protein features of subtype-1 cohort-4 are reflected in other cohort and proteomic features. It can be seen from figure 3 that diffuse gastric cancer with poor survival and MSI gastric cancer with long survival are significantly enriched in RNA1 and RNA2 cluster, respectively. There was no significant difference between rna1 and rna 2 in EOGC and LOGC. It is suggested that the relationship between rna1, rna2 and survival time is similar in young gastric cancer and middle-aged and elderly gastric cancer. Proteomic characteristics, and mRNA characteristics, are not different in EOGC and LOGC, so they are suitable for both EOGC and LOGC.
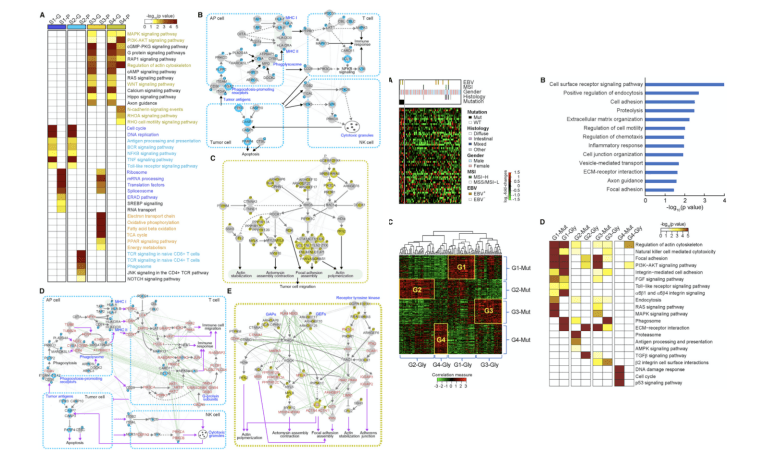 Figure 4. Cellular Networks Associated with Subtypes of EOGCs and Mutation-Glycosylatoin Correlation(Mun DG, et al. 2019)
Figure 4. Cellular Networks Associated with Subtypes of EOGCs and Mutation-Glycosylatoin Correlation(Mun DG, et al. 2019)
Next, from the EOGC cohort network of four subtypes, the author first found the genes and proteins of each subtype. With representative genes and proteins, look for cellular processes through ConsensusPathDB. Among the four common mutant genes, find those with significantly increased levels of glycosylation. Among them, the ARID1A mutation showed a correlation with glycosylated peptide levels in 80 proteins. These up-regulated glycosylation proteins are related to cell migration and immune-related responses. Strongly linked mutant genes and glycosylated proteins are grouped into 4 classes. In the same cellular process, those somatic genes with SNV are associated with glycosylated proteins, so glycosylation data, and similar conclusions to phosphorylation data, provide additional information on mutation-related tumor cell processes information.
The authors found that finding key mutations based on protein levels was no more effective than finding mutations based on high levels of mRNA expression. MRNA-protein correlation can be used to classify proto-oncogenes. Protein abundance, phosphorylation and glycosylation data provide complementary information to mRNA in cancer typing. The mutant gene overlap of EOGC and LOGC is very rare, which indicates that the mutation landscape of the two kinds of gastric cancer is very different. However, there was no significant difference in the expression level of mRNA between the two kinds of gastric cancer. However, there is a significant difference between diffuse and non-diffuse tumors at mRNA level. Since LOGC does not have protein data, it is not possible to compare the levels of protein, phosphorylation and glycosylation between the two types of gastric cancer.
The protein genome analysis in this paper provides signal pathways related to somatic mutations, proto-oncogenes and tumor suppressors. Based on the above findings, early-onset gastric cancer of diffuse type can be divided into two groups: good prognosis group and poor prognosis group, according to (1) whether the mRNA/ protein levels of 4 proto-oncogenes and 2 tumor suppressor genes are up-regulated or down-regulated. (2) according to the patient's mRNA and protein characteristics, whether the patient is assigned to subtype2 or subtype4.
Creative proteomics has an excellent team of experts to provide you with customized multi-group oncology-related experimental design and experimental solutions, so that you can experience the highest quality, the most advanced and the most satisfactory service.Help scientists in the field of cancer research to produce quick results and good results, so as to promote scientific and technological innovation. Creative proteomics, multi-layer assemblage customization service experts to help you with your scientific research!
Reference
1. Mun DG, Bhin J, Kim S, et al. (2019). "Proteogenomic characterization of human early-onset gastric cancer." Cancer cell, 35(1): 111-124.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

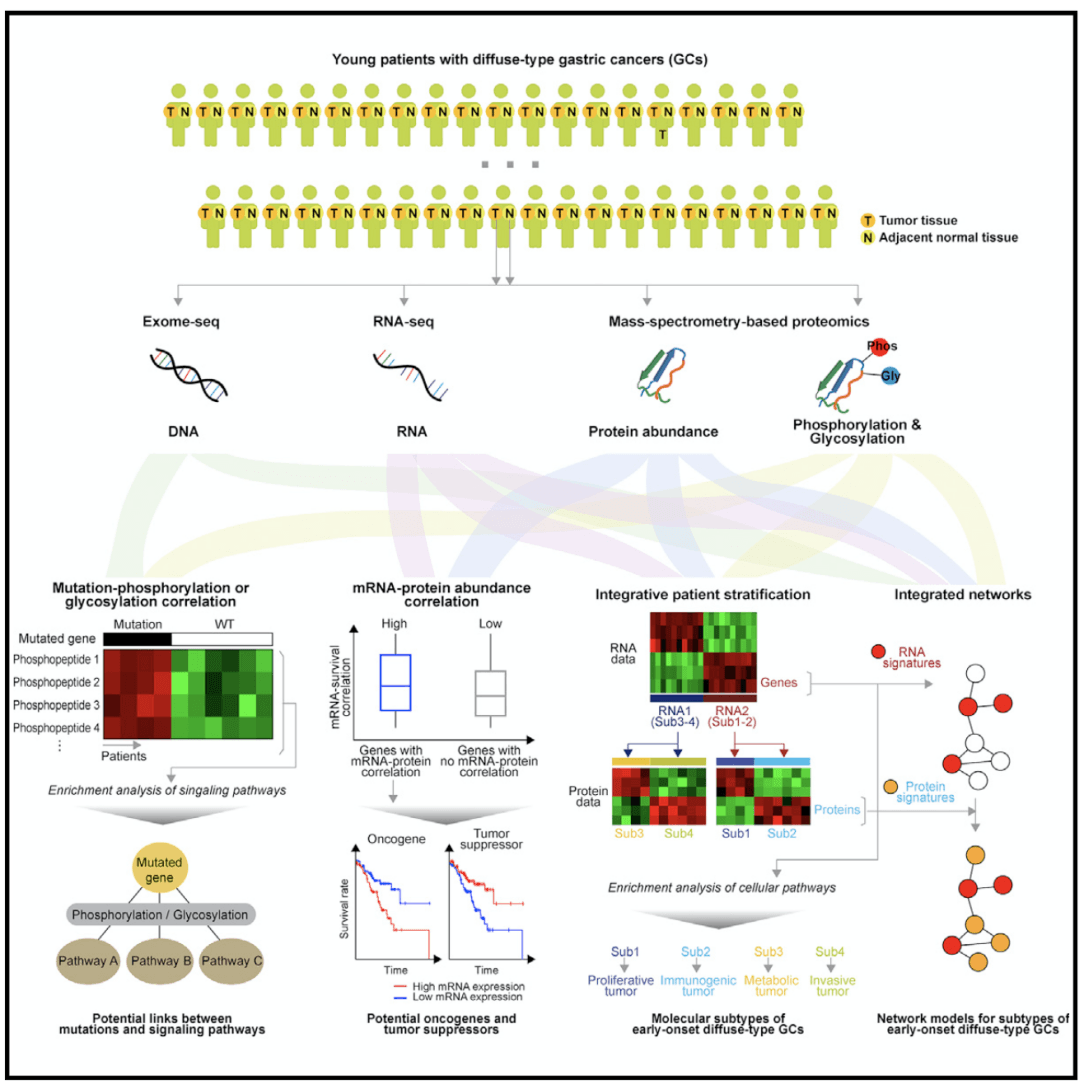
 Figure 1. Proteogenomic Analysis of Nonsynonymous Somatic SNVs .(Mun DG, et al. 2019)
Figure 1. Proteogenomic Analysis of Nonsynonymous Somatic SNVs .(Mun DG, et al. 2019)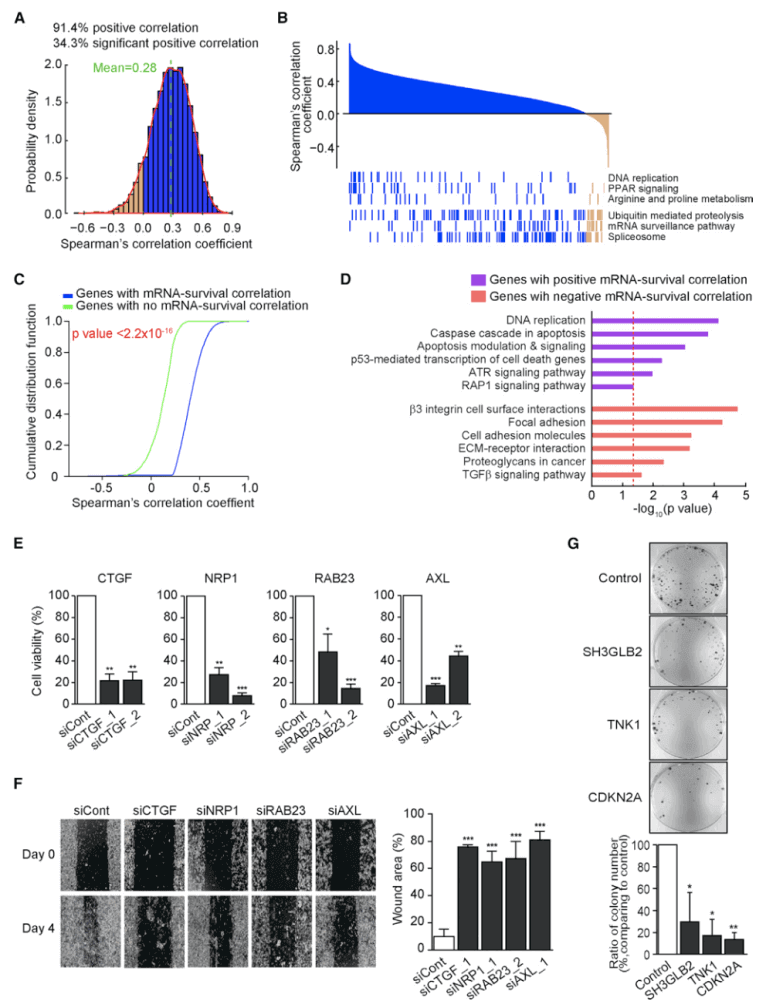
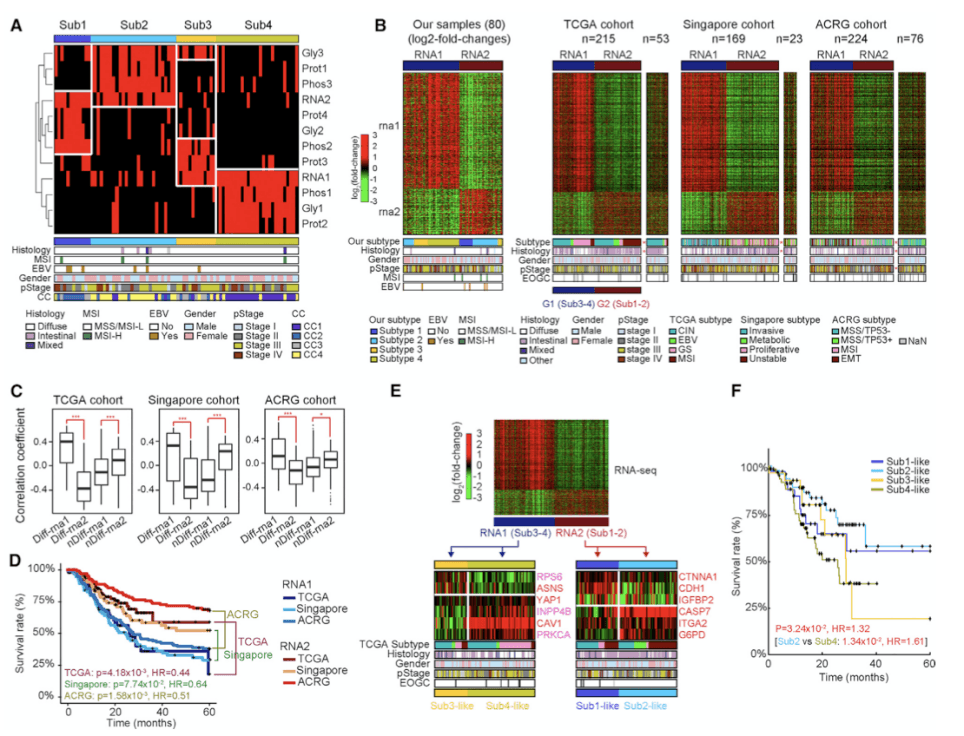
 Figure 4. Cellular Networks Associated with Subtypes of EOGCs and Mutation-Glycosylatoin Correlation(Mun DG, et al. 2019)
Figure 4. Cellular Networks Associated with Subtypes of EOGCs and Mutation-Glycosylatoin Correlation(Mun DG, et al. 2019)