The integrated analysis of tumor transcriptomics and lipidomics refers to the normalization and statistical analysis of batch data from transcriptomes and lipidomics to establish data relationships between different levels of molecules; it also combines functional analysis and metabolic pathways. Analysis of biological functions such as enrichment and molecular interactions, comprehensively analyze the functions and regulatory mechanisms of tumor biomolecules, and finally achieve a comprehensive understanding of the megatrends and directions of biological change. Then propose a molecular biological change mechanism model and screen out key metabolism Pathways, genes, and metabolites for subsequent in-depth experimental analysis and application.
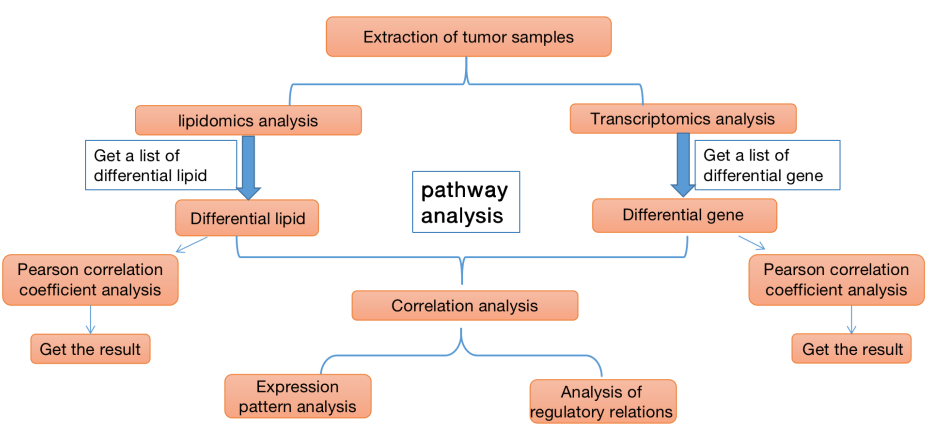 Figure 1. Flow chart of correlation analysis between transcriptomics and lipidomics
Figure 1. Flow chart of correlation analysis between transcriptomics and lipidomics
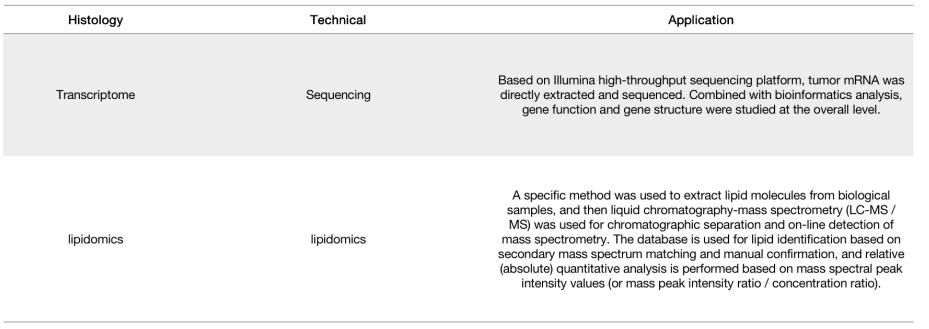
Our advantage

By analyzing different tumor expression levels, we can achieve a full spectrum analysis of tumor genes and lipids, and the mutual verification effect is more obvious.
We explain the molecular regulation-phenotype association mechanism and systematically analyze the tumor biomolecular functions and regulatory mechanisms.
We remove false information from massive data and screen out key tumor metabolic pathways or genes and metabolites for subsequent in-depth experimental analysis and application.
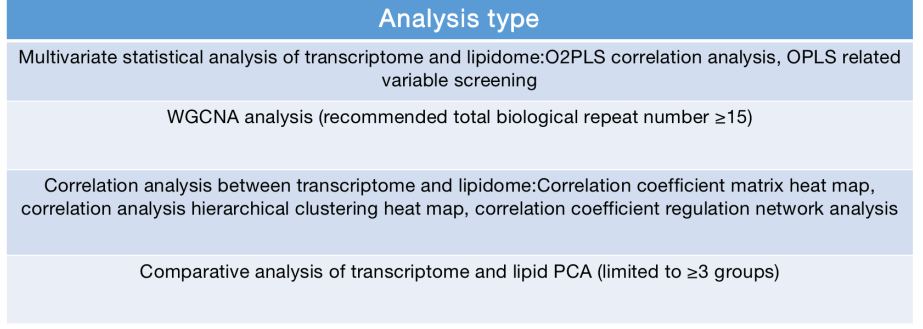
Deliverables: raw sequencing data, pruning and stitching sequences, quality control report results, statistics and bioinformatics reports you specify, visual pictures.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.
Related Services:

 Figure 1. Flow chart of correlation analysis between transcriptomics and lipidomics
Figure 1. Flow chart of correlation analysis between transcriptomics and lipidomics

