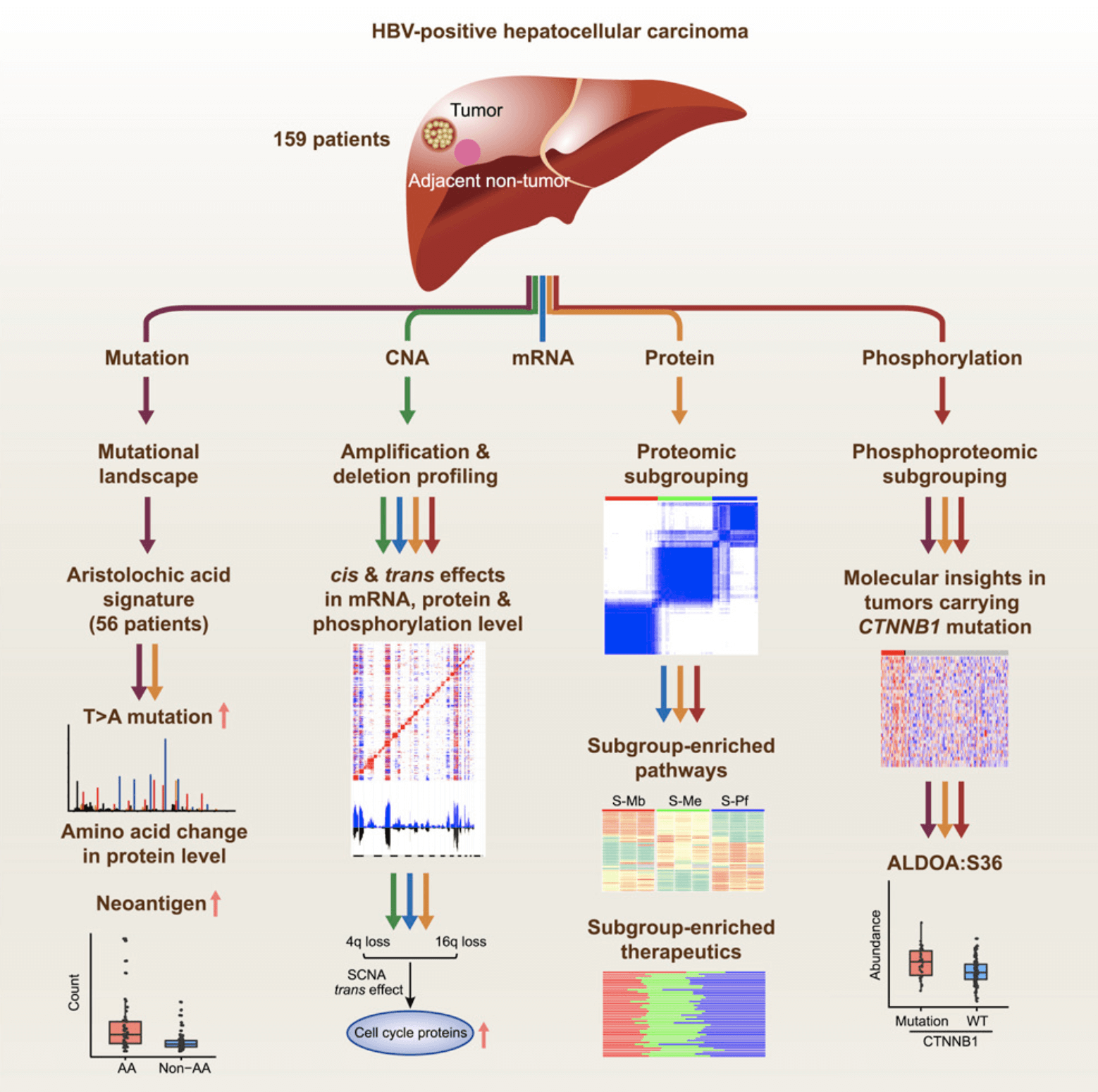
 Figure 1. Study on the protein genome of liver cancer(Gao Q, et al. 2019).
Figure 1. Study on the protein genome of liver cancer(Gao Q, et al. 2019).
Chronic hepatitis B virus (HBV) infection is the most important cause of liver cancer. About 85% of liver cancer patients carry the HBV infection mark. The occurrence of liver cancer is a complex biological process. At the same time, new drug research requires "source innovation", and research on precise molecular typing of liver cancer and potential biomarkers and drug targets related to typing will help the development of personalized medicine for liver cancer. On October 3, 2019, Cell published the latest multi-omics study of liver cancer, including "Integrated Proteogenomic Characterization of HBV-related Hepatocellular Carcinoma". This study carried out protein genomic studies on 159 patients with HBV-infected hepatocellular carcinoma (HCC) and tissue samples from adjacent liver tissues, and analyzed and analyzed gene mutations, copy number variations, gene expression profiles, proteomes and Multi-dimensional data, such as phosphorylated proteome, completely reveals the molecular characteristics of liver cancer, comprehensively analyzes the occurrence and development mechanism of liver cancer, and provides new ideas for accurate typing and personalized treatment of liver cancer, monitoring of curative effects and prognosis. Strategy.
Figure 2. The Workflow of the CHCC-HBV Proteogenomic Study,Quality Assessments for WES, RNA-Seq, and MS Data, Related to STAR Methods(Gao Q, et al. 2019)
First, the study revealed the differences between the mutational spectrum of liver cancer in China and the mutational spectrum of western liver cancer. Aristolochic acid is classified as a class 1 carcinogen by the World Health Organization. The study found that 35% of the samples contained genetic mutation "fingerprints" induced by aristolochic acid. This mutation "fingerprint" has a significant correlation between increased tumor mutation burden and microenvironmental immune tolerance, suggesting the potential value of immunotherapy for this type of liver cancer. Through in-depth analysis of chromosome copy number variations, it was revealed that the deletion of chromosome 4q and 16q will affect cell cycle related proteins and promote the occurrence and development of liver cancer through the trans effect.
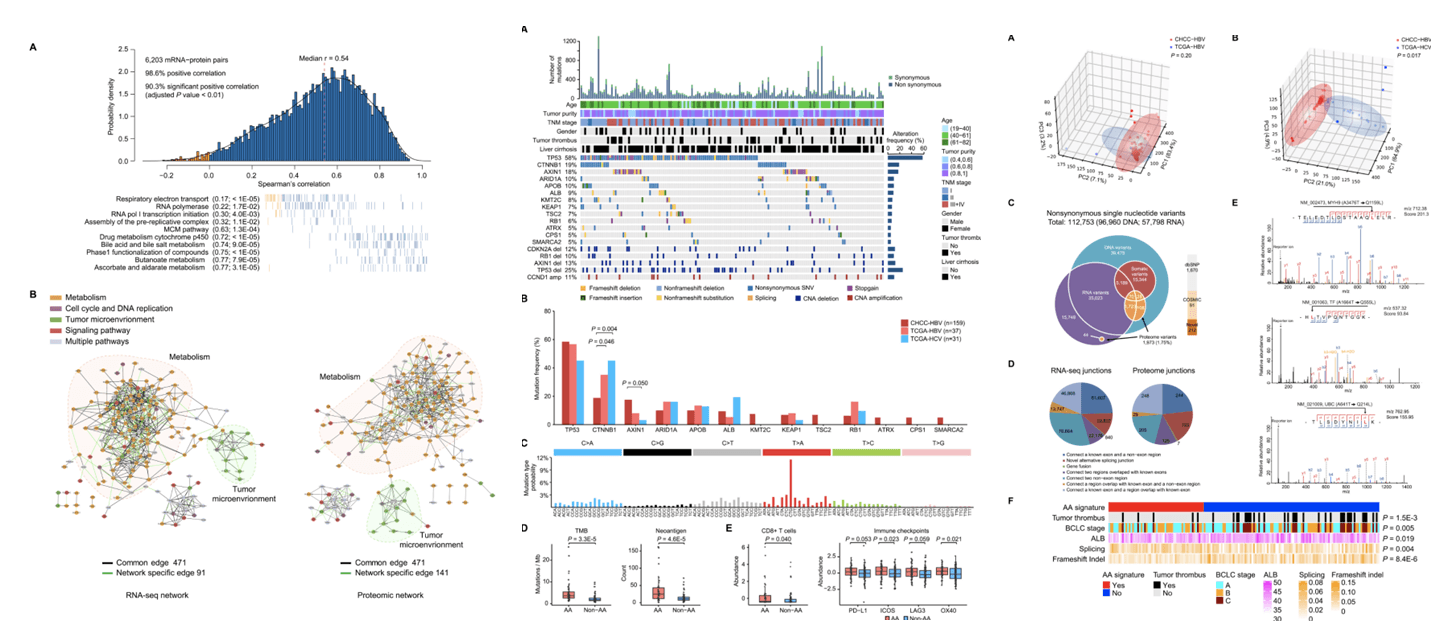 Figure 3. The Overall Correlation, Co-clustering, and Co-expression Network Analyses between mRNA and Protein Data, Related to STAR Methods, WES-Based Mutation Profile of the HBV-Related HCC Cohort, The Mutational Signature Analyses, Direct Effects of Genomic Alterations on Protein Level, and Analysis of AA Mutational Signature in CHCC-HBV(Gao Q, et al. 2019)
Figure 3. The Overall Correlation, Co-clustering, and Co-expression Network Analyses between mRNA and Protein Data, Related to STAR Methods, WES-Based Mutation Profile of the HBV-Related HCC Cohort, The Mutational Signature Analyses, Direct Effects of Genomic Alterations on Protein Level, and Analysis of AA Mutational Signature in CHCC-HBV(Gao Q, et al. 2019)
Second, according to proteomic data, liver cancer patients can be divided into three subtypes, which are metabolic-driven, microenvironmentally dysfunctional, and proliferative-driven. These three subtypes are related to genomic stability, the presence or absence of cancerous plugs, and fetuses. Protein abundance and RB1 and TSC2 gene mutations were significantly related. More importantly, the clinical prognosis of the three subtypes is significantly different, and its prognostic value does not depend on the commonly used TNM staging in the clinic, which is expected to provide guidance for personalized accurate treatment of liver cancer and clinical prognosis judgment.
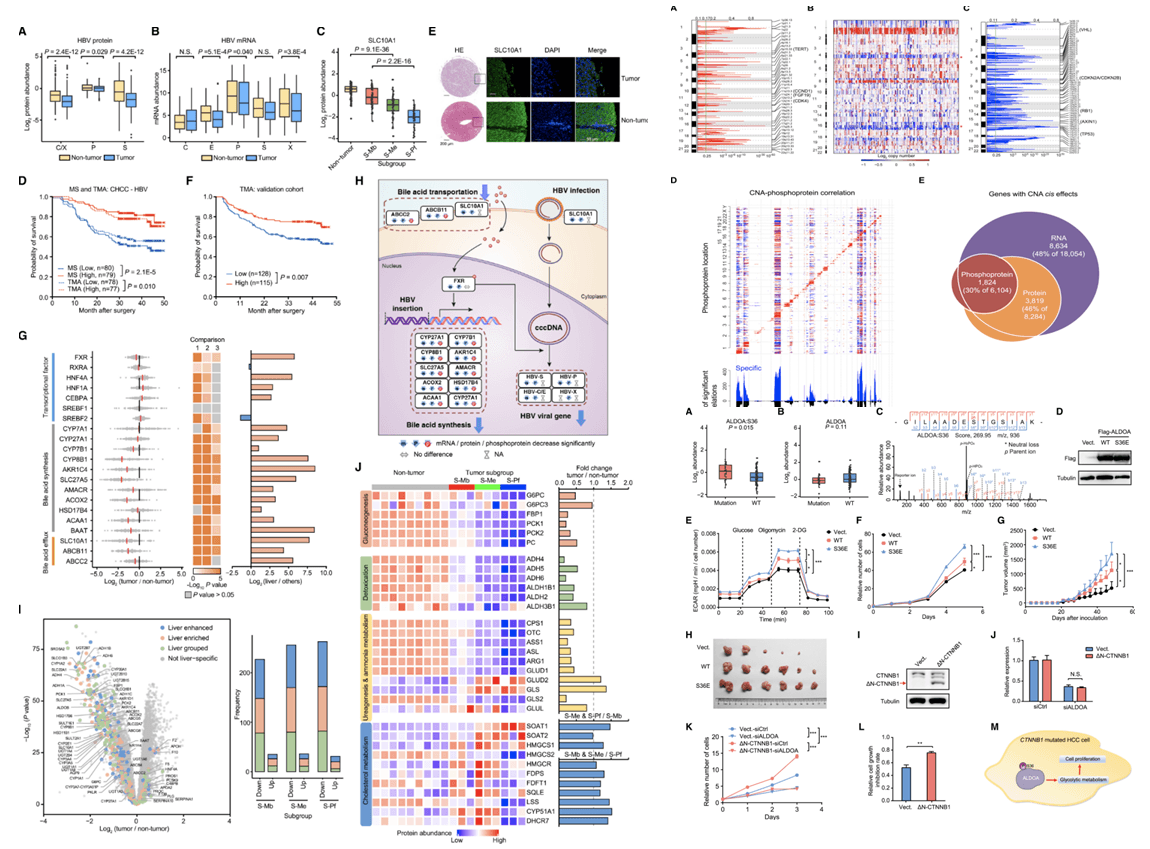 Figure 4. Profiles of Copy-Number Alterations and Correlations of CNA to Phosphoprotein Level in CHCC-HBV Cohort, HBV Receptor and Bile Acid Metabolism Are Downregulated in HBV-Related HCC, ALDOA-Ser36 Phosphorylation Drives HCC Cell Glycolysis and Proliferation(Gao Q, et al. 2019)
Figure 4. Profiles of Copy-Number Alterations and Correlations of CNA to Phosphoprotein Level in CHCC-HBV Cohort, HBV Receptor and Bile Acid Metabolism Are Downregulated in HBV-Related HCC, ALDOA-Ser36 Phosphorylation Drives HCC Cell Glycolysis and Proliferation(Gao Q, et al. 2019)
Finally, the study systematically revealed the important role of metabolic abnormalities in the development of liver cancer. The authors found that 80.3% of liver-specific proteins are down-regulated in tumors and are mainly involved in liver-specific metabolic pathways such as gluconeogenesis, detoxification and urea-ammonia metabolism, while key enzymes for cholesterol metabolism and glutamine metabolism-related proteins Expression is upregulated in tumors. These data reveal that liver-specific functions undergo metabolic reprogramming in HBV-infected HCC tumors. The study also systematically explained the changing trends of various cellular metabolism and signaling pathways in liver cancer from the multi-omics level. Among them, glycolysis and fatty acid metabolism were up-regulated in cancer as a whole, and bile acid metabolism was down-regulated as a whole. As one of the most commonly mutated genes in liver cancer, the differences in protein and phosphorylation between CTNNB1 mutant and wild-type liver cancer are mainly concentrated in metabolic pathways. Analysis showed that the phosphorylation of fructose diphosphate aldolase A (ALDOA) was associated with CTNNB1 mutation. Mechanism studies have shown that ALDOA phosphorylation can enhance the glucose metabolism of liver cancer cells, thereby promoting the growth and tumorigenesis of liver cancer, and the activity of ALDOA is very important for the proliferation of CTNNB1 mutant liver cancer cells. Therefore, targeted regulation of metabolic pathways may play an important role in the prevention and treatment of liver cancer.
This research is the first international multi-level analysis of large-scale clinical samples of HBV-related liver cancer in accordance with the highest standards. The high-quality big data generated will provide support to the majority of liver cancer basic and clinical researchers, thereby effectively promoting the field of liver cancer research. development of.Creative proteomics has an excellent team of experts to provide you with customized multi-group oncology-related experimental design and experimental solutions, so that you can experience the highest quality, the most advanced and the most satisfactory service.Help scientists in the field of cancer research to produce quick results and good results, so as to promote scientific and technological innovation. Creative proteomics, multi-layer assemblage customization service experts to help you with your scientific research!
Reference
1. Gao Q, Zhu H, Dong L, et al. (2019). "Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell", 179(2): 561-577.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.

 Figure 1. Study on the protein genome of liver cancer(Gao Q, et al. 2019).
Figure 1. Study on the protein genome of liver cancer(Gao Q, et al. 2019).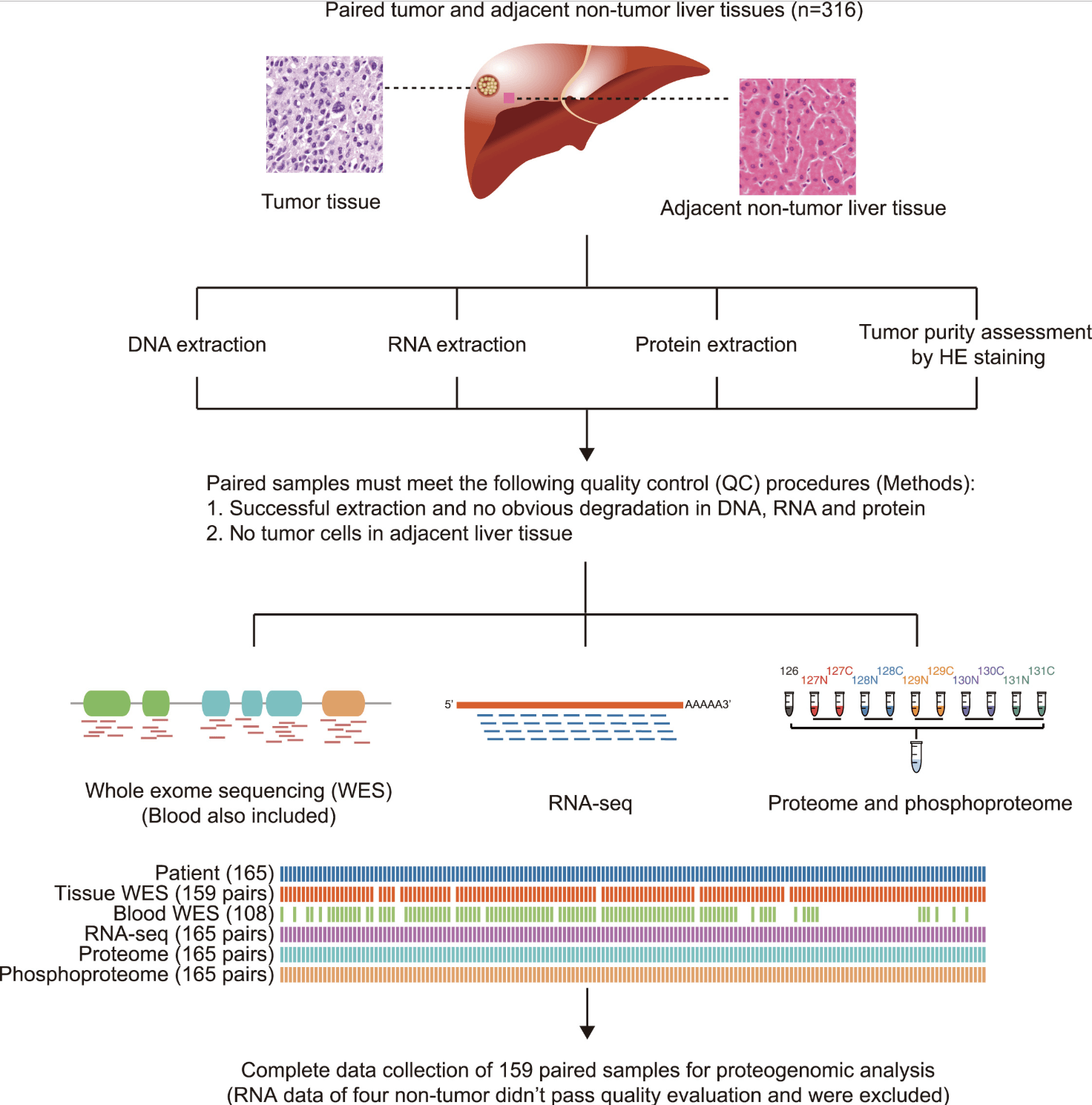
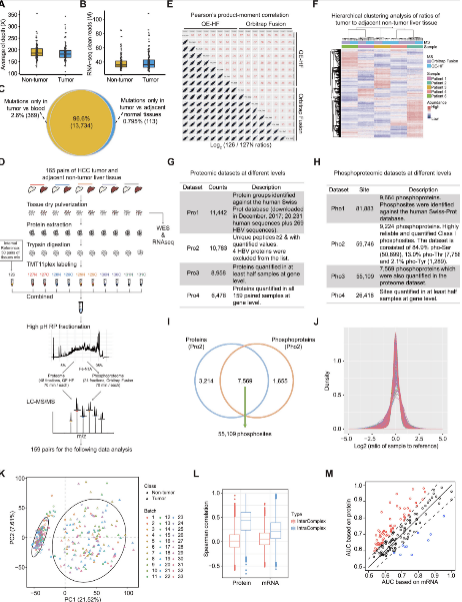
 Figure 3. The Overall Correlation, Co-clustering, and Co-expression Network Analyses between mRNA and Protein Data, Related to STAR Methods, WES-Based Mutation Profile of the HBV-Related HCC Cohort, The Mutational Signature Analyses, Direct Effects of Genomic Alterations on Protein Level, and Analysis of AA Mutational Signature in CHCC-HBV(Gao Q, et al. 2019)
Figure 3. The Overall Correlation, Co-clustering, and Co-expression Network Analyses between mRNA and Protein Data, Related to STAR Methods, WES-Based Mutation Profile of the HBV-Related HCC Cohort, The Mutational Signature Analyses, Direct Effects of Genomic Alterations on Protein Level, and Analysis of AA Mutational Signature in CHCC-HBV(Gao Q, et al. 2019) Figure 4. Profiles of Copy-Number Alterations and Correlations of CNA to Phosphoprotein Level in CHCC-HBV Cohort, HBV Receptor and Bile Acid Metabolism Are Downregulated in HBV-Related HCC, ALDOA-Ser36 Phosphorylation Drives HCC Cell Glycolysis and Proliferation(Gao Q, et al. 2019)
Figure 4. Profiles of Copy-Number Alterations and Correlations of CNA to Phosphoprotein Level in CHCC-HBV Cohort, HBV Receptor and Bile Acid Metabolism Are Downregulated in HBV-Related HCC, ALDOA-Ser36 Phosphorylation Drives HCC Cell Glycolysis and Proliferation(Gao Q, et al. 2019)