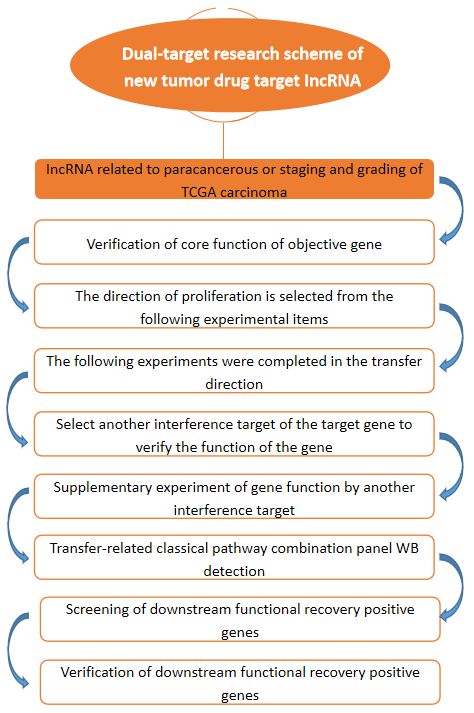
Dual-target Research Scheme of New Tumor Drug Target IncRNA
Based on TCGA big data, we analyzed and verified the new lncRNA functional genes that had not been reported in the studied tumors, and discussed the molecular mechanism of their pathogenesis, so as to provide a preliminary data basis for the determination of follow-up antineoplastic drug targets and biomarkers.
Long-chain non-coding RNA is a non-coding RNA with a length of more than 200bp. More and more studies have shown that lncRNA plays an important role in the occurrence and development of tumors. Through TCGA big data, we analyzed the high expression in cancer tissues or the expression of lncRNA, related to disease staging and grading. We analyzed the candidate lncRNA, that had not been reported in the studied tumors. Using lentivirus-mediated RNAi technology, several candidate lncRNA were RNAi, and the infected cells were counted for 4 to 5 days through the Cellomics high connotation function screening platform, and the target lncRNA, affecting cell proliferation was found. The intracellular localization of target lncRNA was determined by FISH experiment. At least 3 functional experiments of proliferative direction and 2 functional experiments of metastatic direction of the lncRNA in two cell lines were obtained. After interfering with the target lncRNA in a cell line, the transfer-related classical pathway combination panel was verified by WB. According to the results of WB panel, a positive mechanism gene was selected to construct a virus that interfered with or overexpressed the target lncRNA on the cell line, and interfered or overexpressed downstream mechanism genes on the cell line. The changes of cell proliferation ability were observed by HCS scanning for 5 days to verify the functional recovery effect of downstream mechanism genes on target lncRNA, and a positive recovery experiment results of proliferation-related and metastasis-related functional experiments were obtained.
Transfer-related Classical Pathway Combinations Panel
AKT1, p-AKT (Ser473 or Thr308), mTOR, p-mTOR (Ser2448), ERK1/2, p-ERK1/2 (Thr202+Tyr204), P38, p-P38 (T180+Y182), MYC, β-Catenin, p-β-Catenin (S33+S37), NFkB-p65, p-NFkB-p65 (Ser536), CDH1 (E-Cadherin), CDH2 (N-Cadherin), MMP2, MMP9, FN1, VIM, Snail, Slug, TWIST and other 22 genes.
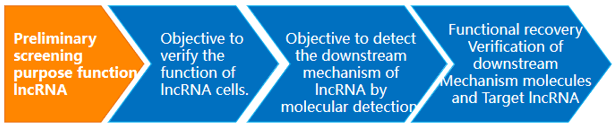
Service Flow
Our Advantage
Mature and reliable experimental analysis platform of Dual-target research scheme of new tumor drug target IncRNA: rich project experience, participation in major projects, high-level cooperation results published.
Based on high-quality data analysis, pre-research and verification of the contents and methods of analysis, it is indeed mature and feasible.
Creative Proteomics leads a rich team that uses the most rigorous experimental solutions and internationally recognized experimental techniques for IncRNA Two-Cell Solutions to obtain your customized information. Use the most accurate experimental results to help you better carry out tumor-related scientific research.
* For Research Use Only. Not for use in the treatment or diagnosis of disease.
Related Services: